FIGURE 2. Mechanisms of nucleosome remodeling by RNA polymerases.
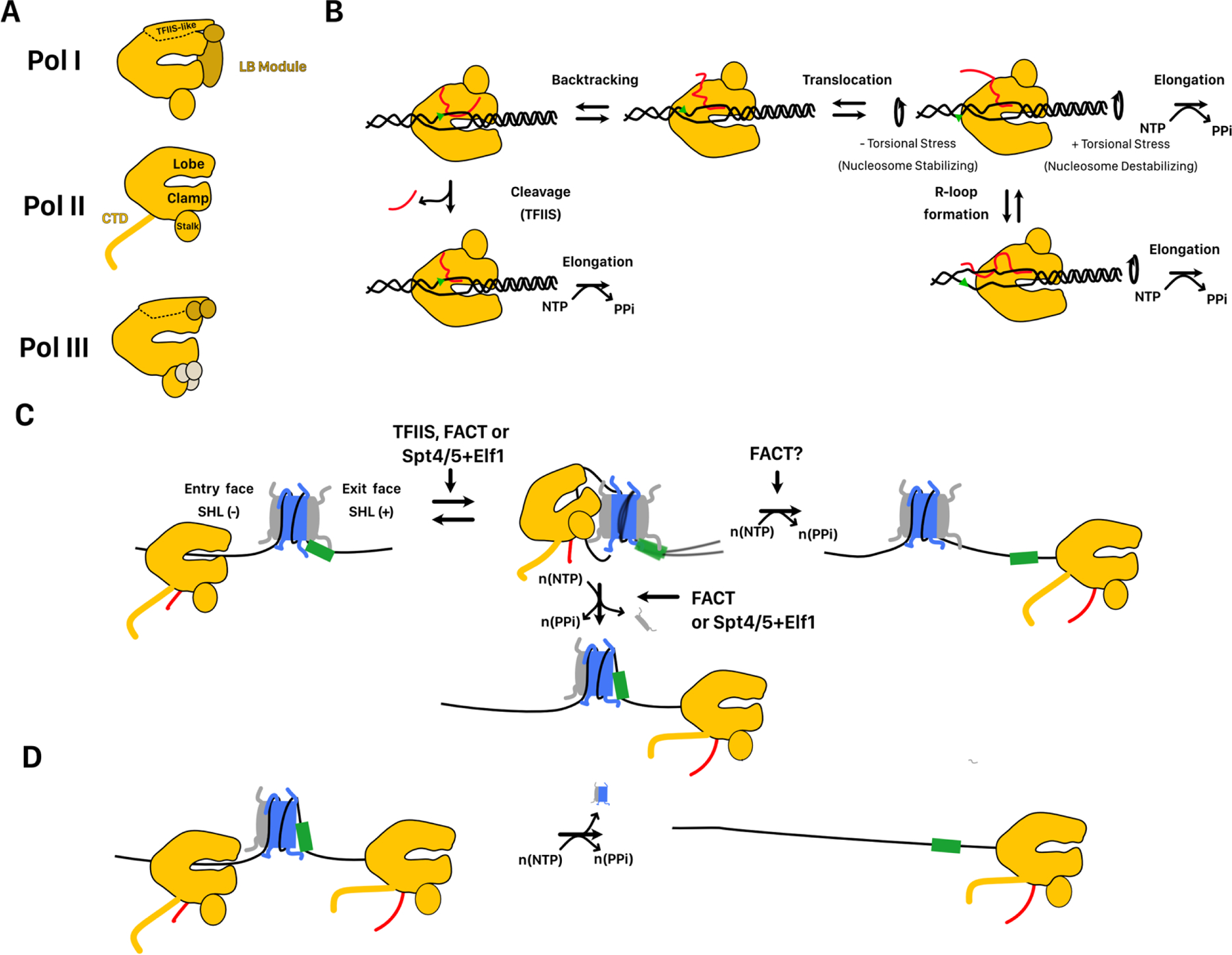
A. Cartoon representations of the three eukaryotic RNA polymerases. The lobe, clamp, and stalk domains that are shared between all three polymerases are labeled on Pol II. The TFIIS like domain of the lobe and the LB module shared by Pol I and III are labeled on Pol I. Finally, the unique CTD of Pol II is also shown. B. Core mechanism of elongation shared by RNA polymerases. The green triangle is used as a reference point for polymerase translocation. Polymerase can either translocate 1 nt forward or pause and backtrack. DNA unwinding is associated with torsional stress ahead of and behind the transcription bubble. Stress behind the bubble can be absorbed by formation of a stable RNA-DNA hybrid called an R-Loop. Backtracked polymerase can be resolved by cleavage of the nascent transcript by TFIIS or the TFIIS-like domains of Pol I and III. Forward elongation is biased by irreversible incorporation of a complementary nucleotide. C. Mechanism of polymerase passage through the nucleosome. The green box is used as a reference location on the DNA. As polymerase translocates into the nucleosome core, it unpeels DNA at the entry side until it reaches the dyad. Once at the dyad, further passage requires DNA unpeeling at the exit side and loosening of histone DNA contacts downstream of the dyad. Further passage is associated with either octamer transfer upstream of its current site or dissociation of an H2A/H2B dimer from the exit side. Elongation factors assist entrance into the nucleosome and may bias the outcome of Pol II passage. D. Polymerase passage through a hexasome by Pol II is associated with complete disassembly of the hexasome.
