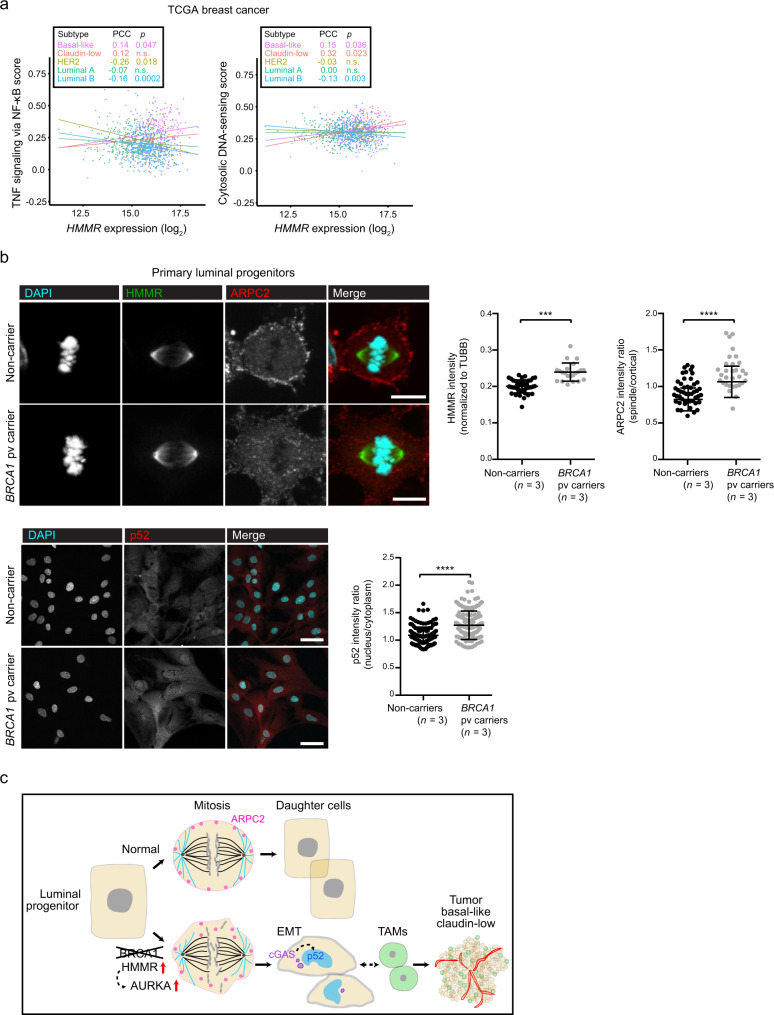
Fig. 7. HMMR-centered perturbations at the basis of breast cancer risk.
a HMMR expression correlation (Pearson’s correlation coefficient (PCC)) with the gene sets “TNF signaling via NF-κB” (left panel) and “Cytosolic DNA-sensing” (KEGG annotation; right panel) across breast cancer subtypes (inset, color-coded; basal-like, n = 213; claudin-low, n = 51; HER2, n = 88; luminal A, n = 234; and luminal B, n = 556). The data corresponded to TCGA primary breast tumor RNA-seq profiles and the PCC p values (or n.s.) are also indicated. b Top and bottom left panels, representative immunofluorescence images of ARPC2, HMMR, and p52 staining in luminal progenitors from healthy non-carrier and carriers of BRCA1 pathological variants (pv). Right panels, quantification: ARPC2; mean ± s.d.; n = 30 cells from non-carriers (n = 3); and n = 30 cells from BRCA1 pv carriers (n = 3); HMMR; mean ± s.d.; n = 31 cells from non-carriers; n = 21 cells from BRCA1 pv carriers; and p52; mean ± s.d.; n = 150 cells in each setting. Two-tailed Student’s unpaired-samples t test; ***p = 0.0002; ****p < 0.0001. Scale bars = 10 μm (ARPC2, HMMR) and 50 μm (p52). c Illustration of the proposed sequence of molecular and cellular events leading to normal luminal progenitor cell division (top sequence) and their alteration in BRCA1-associated breast cancer (bottom sequence). Loss of BRCA1 and HMMR overexpression activate AURKA and reduce cortical retention of ARPC2 during mitosis, disrupting correct chromosome segregation, prompting emergence of micronuclei in daughter cells, which appear imbalanced in size and have undergone EMT as a consequence. Then, micronuclei-induced cGAS signaling activates non-canonical NF-κB, which can facilitate recruitment of TAMs that enable initial tumorigenesis. As a consequence, developed tumors are predicted to show basal-like and claudin-low features with high vascularization and immune cell infiltration.