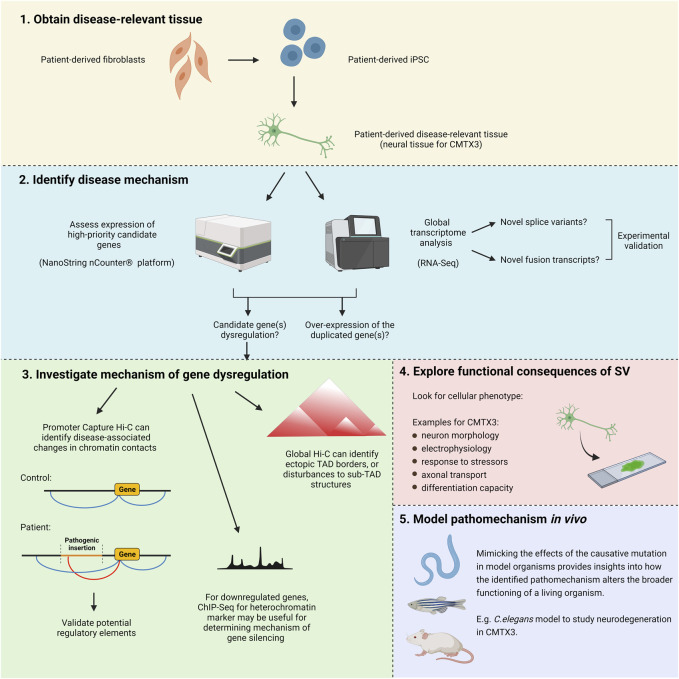
FIGURE 4.
Experimental approach for uncovering the pathogenic mechanism of CMTX3 and the other diseases associated with insertions(+/-deletions) at Xq27.1. (1) Induced pluripotent stem cells retaining the patient genetic background can be used to generate patient-derived disease-relevant tissue for downstream analysis. (2) The expression level of high-priority candidate genes can be specifically assessed through NanoString nCounter® analysis. Global transcriptomic analysis can be carried out using RNA-Seq. In addition to the predominant hypothesis of gene dysregulation, this approach directly addresses alternative pathomechanisms previously hypothesised to underly these diseases, such as abnormal splice variants, aberrant fusion transcripts, and overexpression of the duplicated genes. (3) If gene dysregulation is observed, experiments can help to uncover the underlying mechanism. Promoter Capture Hi-C can identify alterations in long-range regulatory interactions. Global Hi-C can identify ectopic TAD boundaries that might be introduced by the insertions, as well as changes to tissue-specific sub-TAD organisation. If the disease mechanism involves gene repression, ChIP-Seq for a heterochromatin marker (H3K9me3) can identify whether this arises from mutation-induced alterations to the local chromatin environment rather than altered regulatory interactions (Laugsch et al., 2019). (4) A range of functional tests can be performed to look for a cellular phenotype in patient samples. (5) Potential pathomechanisms can be studied in vivo using model organisms. Image created with BioRender.com.