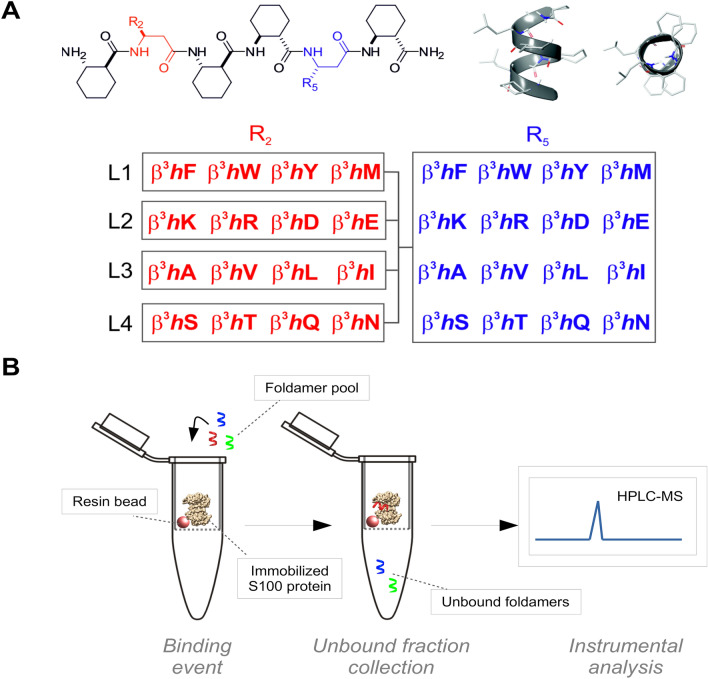
Figure 1.
The methodology of the high-throughput (HTP) holdup (HU) assay. (Panel A) General sequence of the foldamer library members and structure of H14 helix in side and plan view generated manually by Schrödinger Maestro 11.7 molecular modelling software. The 256-membered foldamer library was divided into 4 sublibraries (L1–L4) based on the general characteristic of the second amino acid (labelled with red) in the sequence. Each sublibrary consists of 64 individuals (R2: four different amino acids, R5: sixteen different amino acids). These four sublibraries (L1–L4) are aromatic, charged, aliphatic, and polar, respectively. (Panel B) His-tagged S100 proteins immobilized on Co2+-resin (left panel) are incubated with the H14 foldamer library (256 members). The unbound fraction (flow-through) is recovered (middle panel) and the flow-through fractions are analyzed by LC–MS.