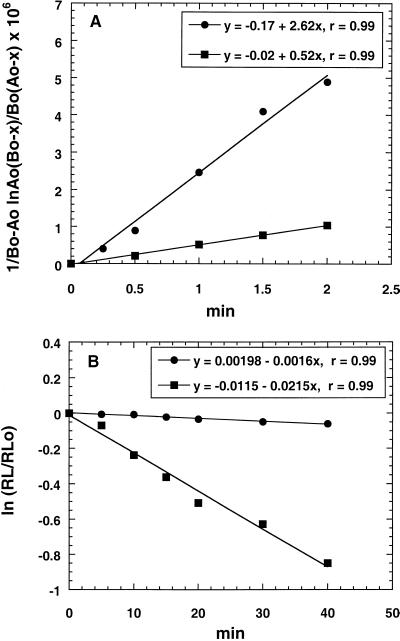
FIG. 2.
Kinetic analysis of drug association and dissociation in S. pneumoniae 2486. (A) Drug on rate determinations. Cells (1.0 × 109 CFU/ml) in Miller's M9 medium were incubated at 23°C with either 0.2 μg of [14C]erythromycin (squares) or [14C]ABT-773 (circles) per ml for short intervals up to 2 min. Labeled cells were processed as outlined in Materials and Methods. Influx of label was plotted as 1/(Bo − Ao) [ln Ao(Bo − x)/Bo(Ao − x)] versus t, where Ao is concentration of free ribosomes, Bo is concentration of free drug at time zero, and x is cell-associated drug at time t. The slope is equal to k1. (B) Drug off rate determinations. Cells (1.2 × 109 to 1.5 × 109 CFU/ml) in Miller's M9 medium were incubated at 23°C with 0.2 μg of either [14C]erythromycin (squares) or [14C]ABT-773 (circles) per ml for 30 min (saturation of binding sites). Labeled cells were harvested and suspended in fresh medium containing 0.2 μg of unlabeled ABT-773 per ml. Efflux of label at 23°C was monitored for 40 min and plotted as ln(RL/RLo) versus t, where RL is cell-associated label at time t, RLo is cell-associated label at zero time, and the slope is k−1.