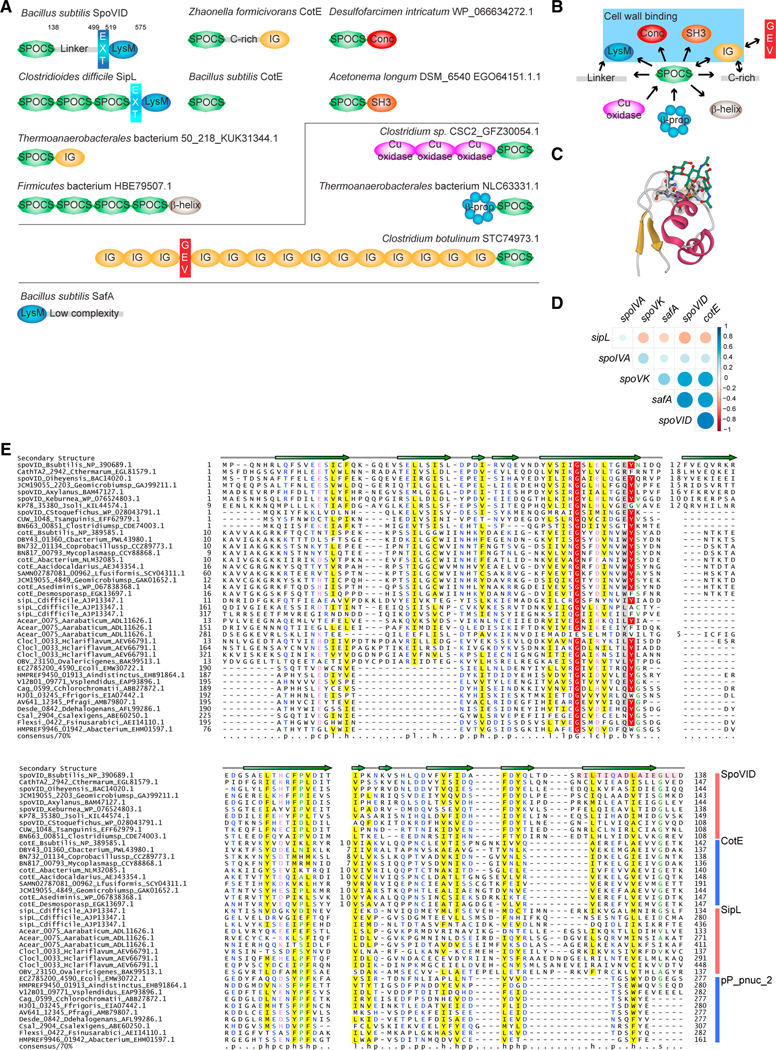
Figure 2. SpoVID domain architecture and phylogeny reveal a superfamily of coat assembly proteins.
(A) Domain architectures of SPOCS (SpoVID-CotE-SipL) domain-containing proteins. Domain architectures are labeled with organism name and a representative gene name (whenever a known gene name is present) or accession. GEV, GEVED; conc, Concanavalin-like; β-prop, beta propeller; EXT, N-terminal extension in LysM domains found in SpoVID and SipL (difference in color denotes a difference in the nature of the extension). Lines delineate proteins harboring N-terminal or C-terminal SPOCS domain(s). For B. subtilis, numbers indicate residue positions that mark C-terminal boundaries of each domain. Also depicted is B. subtilis SafA, a sporulation protein harboring a LysM domain that interacts with SpoVID but does not contain a SPOCS domain. Domains are not drawn to scale.
(B) A simple network is depicted to summarize observed domain arrangements. Arrows represent amino acid sequence direction, from N terminus to C terminus.Blue box indicates domains associated with SPOCS domain that have been implicated in binding peptidoglycan.
(C) The first LysM domain of the NlpC/P60 family peptidoglycan D-L endopeptidase bound to three GlcNAc moieties (green) (PDB: 4UZ3). The threonine facing the reader (circled in yellow) is the equivalent of T532 in SpoVID. The remaining conserved residues of the binding pocket are shown in ball and stick and Gaussian volume representations. Gold, β-strands; maroon, α-helices; green, Glc-Nac moieties; red, O; blue, N; black, C.
(D) Correlation matrix of SpoIVA, SpoVK, SafA, and SPOCS domain-containing proteins SpoVID, CotE, and sipL/DUF3794. Presence or absence of the each of these protein in a given genome is correlated. Positive correlations are displayed in shades of blue and negative correlations in shades of red color. Color intensity and the size of the circle are proportional to the correlation coefficient.
(E) Sequence alignment of the SPOCS domain. Representative set of SPOCS domains from SpoVID, CotE, SipL, and pp_pnuc_2 families is shown in multiple sequence alignment. The encasement region of SpoVID is shown with a pink box. The predicted secondary structure is shown above the alignment with the arrows representing β-strands. Inserts are shown with numbers. The 70% consensus shown below the alignment was derived for the SPOCS domain using the following amino acid classes: hydrophobic (h: ALICVMYFW, yellow), small (s: ACDGNPSTV, green), polar (p: CDEHKNQRST, blue) and its charged subset (c: DEHKR, pink), and big (b: FILMQRWYEK; gray). Numbers at the beginning and end are the start and end of the sequence for the SPOCS domain in a given protein. Numbers in the middle are the length of nonconserved inserts. Aacidocaldarius, Alicyclobacillus acidocaldarius; Aarabaticum, Acetohalobium arabaticum; Abacterium, Acetobacteraceae AT-5844; Abacterium, Acholeplasmataceae bacterium; Aindistinctus, Alistipes indistinctus; Asediminis, Amphibacillus sediminis; Axylanus, Amphibacillus xylanus; Bsubtilis, Bacillus subtilis; Cstoquefichus, Candidatus stoquefichus massiliensis; Cbacterium, Clostridiaceae bacterium; Cchlorochromatii, Chlorobium chlorochromatii; Cdifficile, Clostridioides difficile; Clostridiumsp, Clostridium sp. CAG:451; Coprobacillussp, Coprobacillus sp. CAG:605; Csalexigens, Chromohalobacter salexigens; Cthermarum, Caldalkalibacillus thermarum; Ddehalogenans, Desulfitobacterium dehalogenans; E. coli, Escherichia coli; Ffrigoris, Flavobacterium frigoris; Fsinusarabici, Flexistipes sinusarabici; Geomicrobiumsp, Geomicrobium sp. JCM19055; Hclariflavum, Hungateiclostridium clariflavum DSM19732; Jsoli, Jeotgalibacillus soli; Keburnea, Kroppenstedtia eburnea; Lfusiformis, Lysinibacillus fusiformis; Mycoplasmasp, Mycoplasma sp. CAG:956; Oiheyensis, Oceanobacillus iheyensis; Ovalericigenes, Oscillibacter valericigenes; Pfragi, Pseudomonas fragi; Tsanguinis, Turicibacter sanguinis; Vsplendidus, Vibrio splendidus.