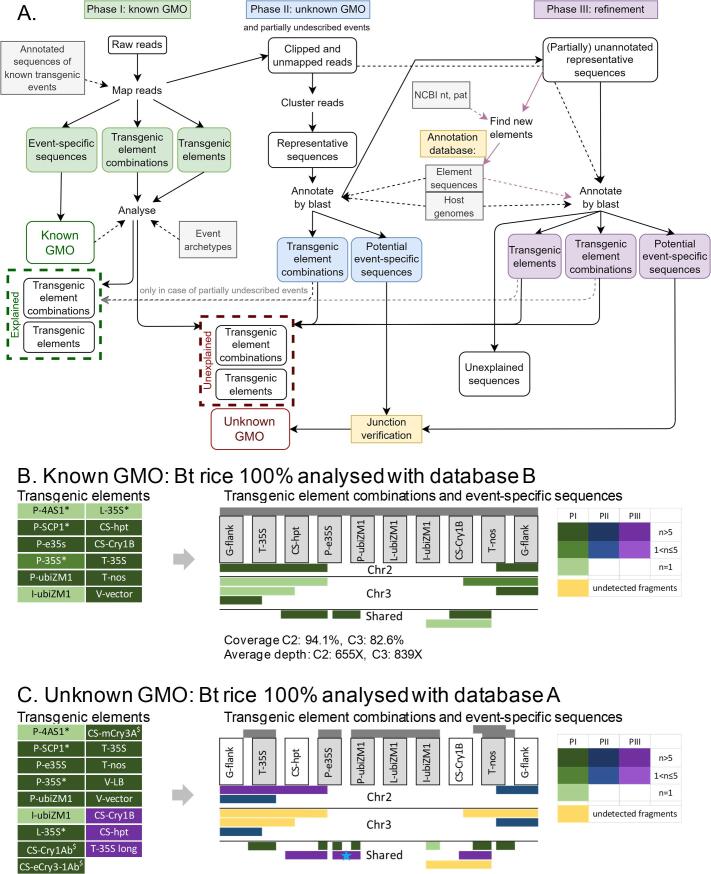
Fig. 1.
Data analysis workflow for detection of known and unknown GMO in DNA walking data. (A) Panel A shows the data analysis workflow. In the first phase, long-read sequencing data is aligned to the annotated sequences of known GMOs from the Nexplorer database, listing all the elements, element combinations and event-specific sequences such as junctions between the transgenic insert and host plant genome that are represented in the sample. Based on the observed event-specific sequences, a list is made of all the events whose presence in a sample is confirmed (Known GMO). The events archetypes (which describe the transgenic elements that are located on an insert in the correct order) are consulted in the database to verify whether all of the observed elements and element combinations can be explained by the presence of detected events. Unexplained elements and element combinations can originate either from known GMOs for which no event-specific sequences were detected in the sample, or from unknown GMOs. During the second phase, which aims at the detection of unknown GMOs, all reads that map partially (clipped reads) and unmapped reads are collected, clustered and annotated by blasting the representative reads of the clusters against the annotation database, consisting of host plant representative genomic sequences (Host genomes), sequences of transgenic elements extracted from the Nexplorer database prior to each analysis and clustered to remove redundant elements (Element sequences) and contaminating sequences such as PacBio internal control sequence and sequences of microorganisms (Contaminants). This phase is performed in case the sample contains clipped or unmapped reads, independent of whether unexplained transgenic elements were observed to avoid potential masking of unknown GMOs by known GMOs with the same elements. Blast results are visualized, allowing to quickly identify the element combinations and potential event-specific sequences that were not observed in phase I, and thus possibly belong to unknown GMOs. The sequences containing potential transgenic junction regions are redirected to the junction verification step. Besides the unknown GMO, phase II allows to reconstruct sequences of known events for which the insert sequence is only partially represented in the database. In the third, optional phase, the obtained annotation results are refined. Representative sequences that were not, or only partially annotated in the previous step are blasted to NCBI nt and pat (patent) databases, and the annotation database is extended with the newly identified transgenic elements and contaminating sequences. The annotation step is repeated with the newly extended annotation database which allows to detect most transgenic elements and element combinations that are not included in the database, and to describe junction regions if the insert portion of the junction does not contain any elements and consists of e.g. transgenic vector sequence. Exceptionally, clipped reads of interest can be extracted and annotated individually, e.g. when all reads containing unexplained transgenic element(s) were discarded during clustering. (B, C) Panels B and C demonstrate the result obtained by applying the described data analysis methodology on rice grain sample containing Bt rice at 100%. The analysis was performed using version B of the database that contains Bt rice sequences (panel B) to represent a scenario where a sample contains a known GMO at 100%, and using version A of the database lacking Bt rice (panel C), representing a scenario where the sample contains an unknown GMO at 100%. Transgenic expression cassettes are shown schematically, elements are not drawn to scale. C2 and C3 stand for chromosomes 2 and 3 respectively. Grey bars above the cassettes indicate which elements and element combinations are present in each version of the database. Elements that are represented in the database that is used for the analysis are displayed in grey boxes. Colored bars below the expression cassette show the longest amplicons that were observed for the first time during phase I, II and III (PI, PII, PIII) of the analysis as described in A. The green/blue/purple colour gradient according to the color legend shows the number of reads (n) representing a given amplicon. Fragments that are known to be present in the sample but that could not be detected in the current analysis are colored yellow. Fragments that were manually reconstructed from reads (instead of clusters) during phase III are marked by a blue star. * Transgenic elements containing homologous regions with P-e35S. $ Transgenic elements containing homologous regions with CS-Cry1B. Transgenic elements are described in Supplementary File 3.