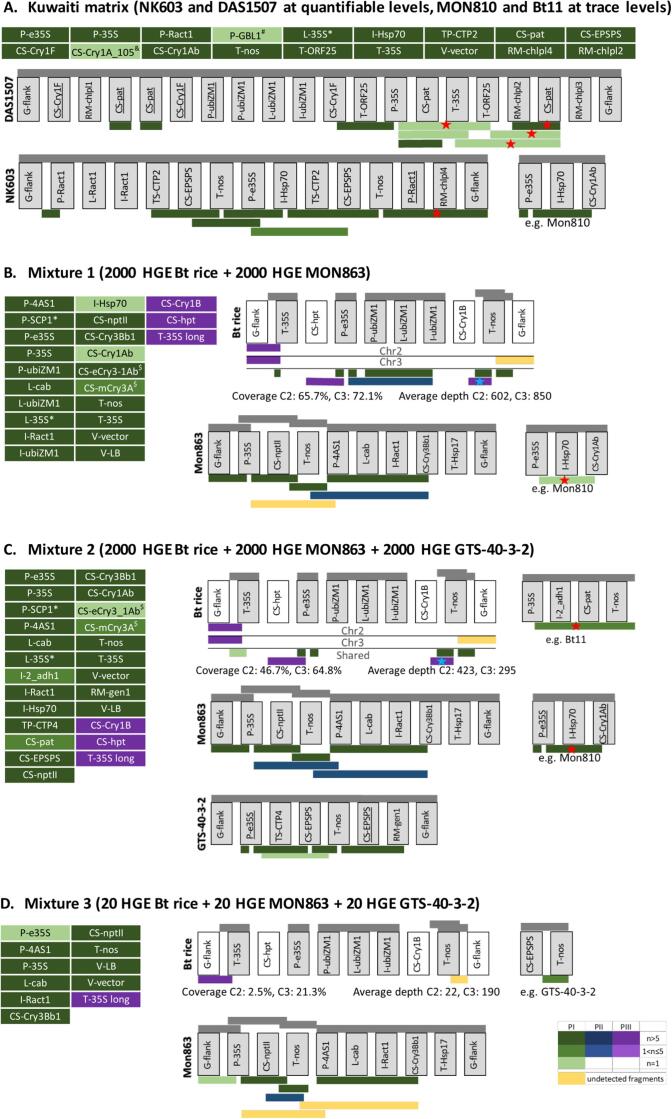
Fig. 4.
Characterization of samples containing mixtures of known and unknown GM events. The data analysis workflow described in Fig. 1A was applied on long-read sequencing data of DNA walking amplicons obtained from rice grain material containing a mixture of (A) known, or (B, C, D) known, and unknown events. Transgenic expression cassettes are shown schematically, and the same cassette is used to represent both transgenic inserts of Bt rice (C2 and C3, corresponding to inserts in chromosome 2 and 3 from rice genome respectively). Grey bars above the cassettes indicate schematically which elements and element combinations are present in the database. Elements that are represented in the database are indicated in grey colored boxes. Truncated transgenic elements are underlined. Colored bars below the expression cassette show the longest amplicons that were observed for the first time during phase I, II and III (PI, PII, PIII) of the analysis as described in Fig. 1A. The green/blue/purple colour gradient according to the color legend shows the number of reads (n) representing a given amplicon. Fragments that are known to be present in the sample but that could not be detected in the current analysis are colored yellow. Fragments that were manually reconstructed from reads (instead of clusters) during phase III are marked by a blue star. Fragments that were not found previously by Fraiture et al (Fraiture et al., 2017) are marked by a red star. *Transgenic elements containing homologous regions with P-35S, P-e35S and P-4AS1. $Transgenic elements containing homologous regions with CS-Cry1B. &Transgenic elements containing homologous regions with CS-Cry1Ab. #Transgenic elements likely originating from the host plant genome. Transgenic elements are described in Supplementary File 3.