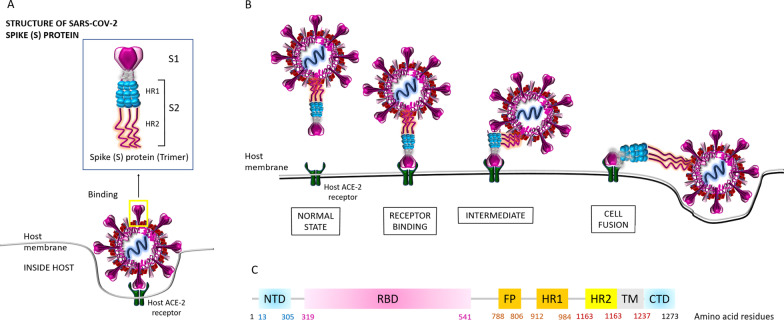
Fig. 2.
SARS-CoV-2 spike (S) protein structure. A The binding mechanism of the host ACE-2 receptor by the SARS-CoV-2 spike (S) protein which has 1273 amino acid residues (180–200 kDa) has been shown. S protein trimers are the crown-like halo structures surrounding the viral particle. The S1 and S2 subunits form the bulbous head and stalk region (Noor and Maniha 2020). B Binding of the host ACE-2 receptor with the SARS-CoV-2 spike protein leading toward the viral fusion within the host has been shown. Indeed, different conformations of the spike (S) RBD domain in opened and closed states function in this mechanism which has been elaborately discussed by Huang et al. (2020) (not shown in this diagram). C Amino acid alignment within the SARS-CoV-2 Spike (S) protein. The S protein contains (1) the extracellular N-terminus domain (amino acids 1–13), (2) the S1 subunit (14–685 residues), and (3) the S2 subunit (686–1273 residues): the fusion peptide (FP) (788–806 residues) plus the heptapeptide repeat sequence 1 (HR1) (912–984 residues) and HR2 (1163–1213 residues); a transmembrane (TM) domain (1213–1237 residues) across the viral membrane, one region for receptor binding and one for membrane fusion; and finally, an intracellular C-terminal domain (CTD/ cytoplasm domain (1237–1273 residues) (Noor and Maniha 2020). As stated earlier, the RBD situated in the S1 subunit binds to the cell host ACE2 receptor (Kahn and McIntosh 2005). Besides, FP is actually a short segment of 15–20 conserved hydrophobic amino acid residues (mostly glycine alanine), which mediates the anchoring of the target membrane when the S protein adopts the conformation (Huang et al. 2020). Moreover, this is noteworthy that the targeting the heptad repeat (HR) has attracted the greatest interest in therapeutic drug discovery so far (Noor and Maniha 2020)