Figure 2.
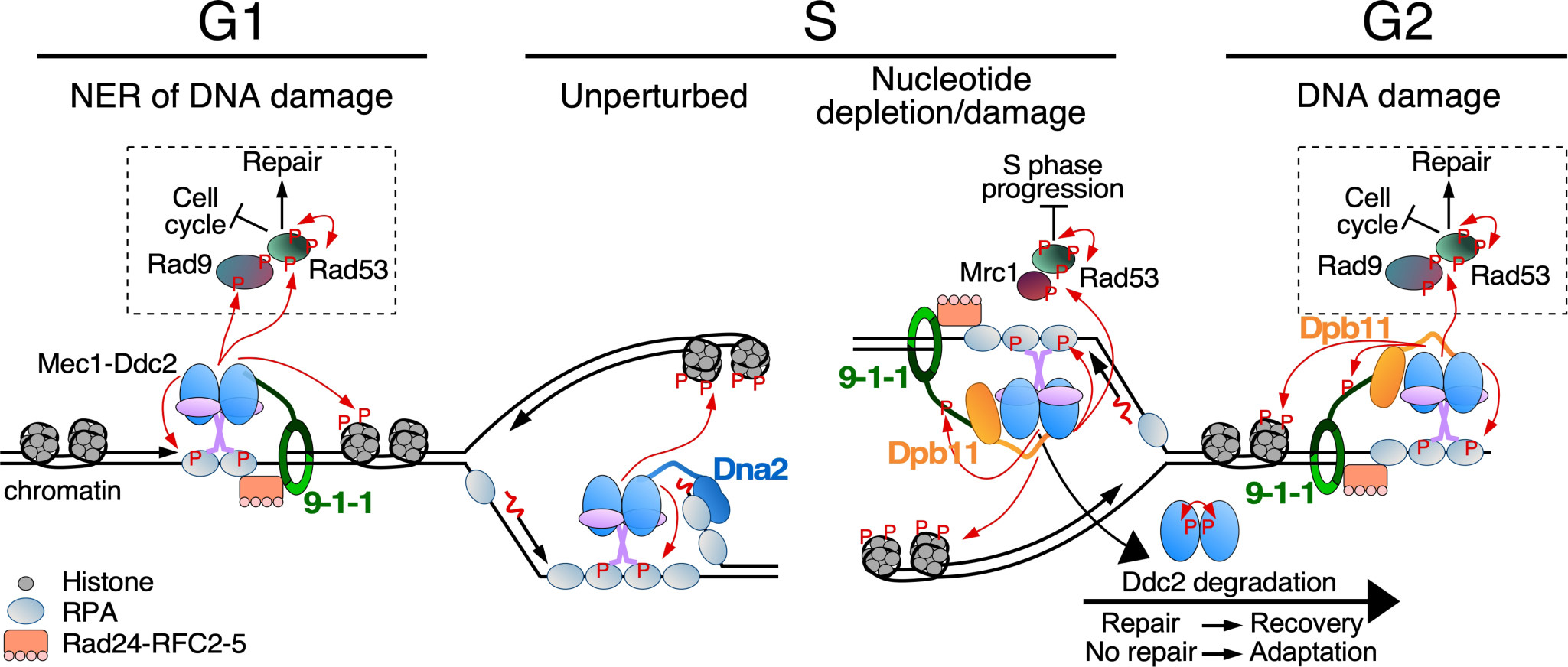
Overview of Mec1-initiated cell cycle checkpoints. Indicated are Mec1 activity (1) during G1 at RPA-coated ssDNA formed as a result of nucleotide excision repair (NER); (2) during S at an unperturbed DNA replication fork or (3) at a stalled fork due to nucleotide depletion or DNA damage; (4) during G2 in response to DNA damage. Mec1-Ddc2 binds RPA-ssDNA and localizes together with Mec1 cell cycle specific activators at the sites of lesions. The 9–1-1 checkpoint clamp is loaded by the Rad24-RFC2–5 clamp loader (human Rad17-RFC2–5) onto gapped DNA, and stimulates Mec1. Phosphorylation of 9–1-1 in S/G2 in response to DNA damage promotes recruitment of Dpb11. Among the many targets of Mec1/ATR is histone H2A (a hallmark of DNA damage), the effector kinase Rad53, and mediator scaffold proteins Rad9 (for DNA damage) or Mrc1 (for replication stress). These scaffold proteins promote auto-hyperphosphorylation of Rad53, promoting downstream pathways including cell cycle arrest and DNA repair. During unperturbed DNA replication, Mec1 may be localized to RPA-coated ssDNA on the lagging strand, and be activated by the nuclease-helicase Dna2, which localizes to DNA flaps. Mec1 auto-phosphorylation and Ddc2 degradation contribute to checkpoint inactivation and adaptation.
