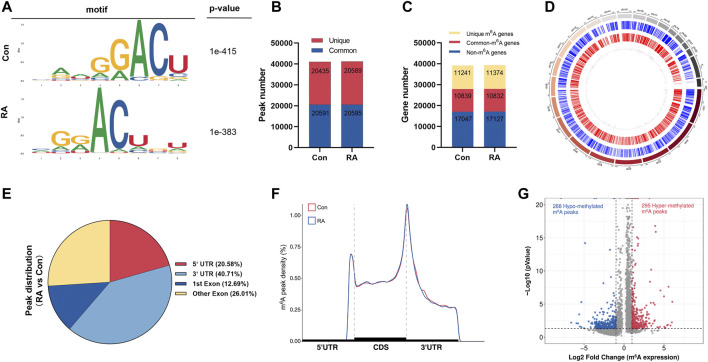
FIGURE 1.
Characteristics of m6A methylation in RA PBMCs. (A) Recognition of the top consensus motif from m6A peaks determined in PBMCs from the RA and Con groups. (B) Number of m6A peaks recognized by m6A-seq in the RA and Con groups. (C) Summary of the m6A-altered genes identified using m6A-seq. (D) Distribution patterns of m6A peaks in various chromosomes. (E) m6A modification distribution in different gene contexts. (F) Accumulation of m6A peaks across transcripts fallen into5′-UTR, CDS, and 3′-UTR. (G) Recognition of the abundance of hypermethylated and hypomethylated m6A peaks in the RA PBMCs relative to that of the Con group. m6A, N 6 -methyladenosine; RA, rheumatoid arthritis; PBMCs, peripheral blood mononuclear cells; and Con, control group.