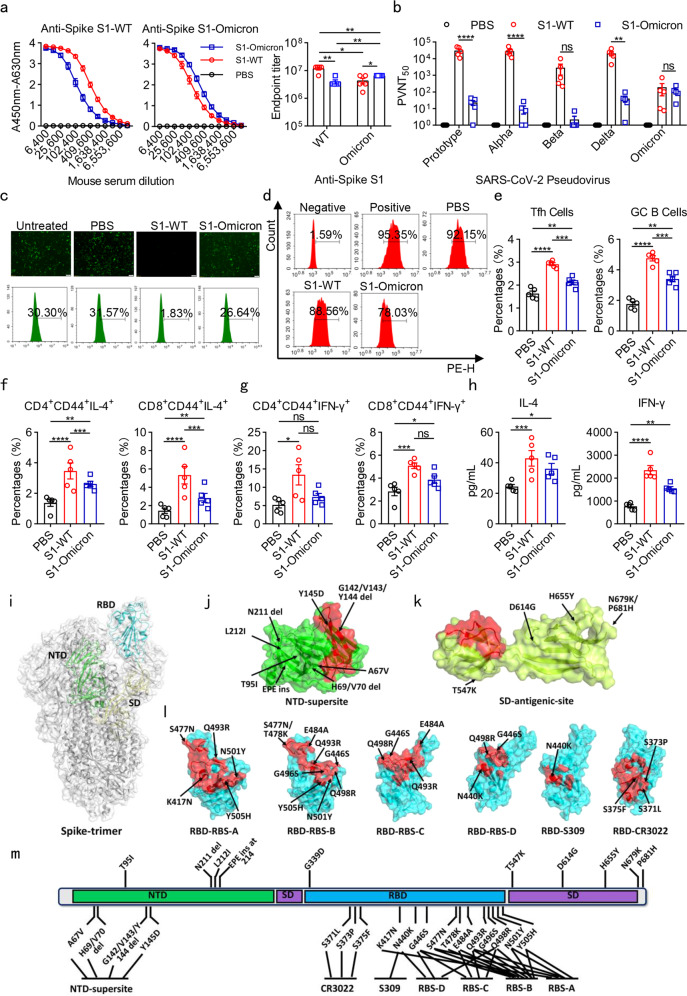
Fig. 1.
Recombinant Omicron S1 protein vaccine induced weak humoral and cellular immunity in mice. a Estimation of the binding ability of sera from immunized with PBS, S1-WT, and S1-Omicron proteins to S1-WT (left) and S1-Omicron (middle)-coated antigens by ELISA. The absorbance was read at 450–630 nm. Antibody titers of S1-WT or S1-Omicron IgG (Right) in sera collected from NIH mice immunized with S1-WT or S1-Omicron vaccine. b PVNT50 of the immune sera against prototype, Alpha, Beta, Delta, and Omicron pseudoviruses. The value was defined as the inverse dilution that achieved 50% neutralization and calculated by GraphPad Prism 8.0. c The blocking effects of sera from immunized mice on the infection of SARS-CoV-2 EGFP/Luciferase-expressing pseudovirus into 293T/ACE2 cells. Immune sera samples were diluted at 1:90. d Representative flow cytometry showing the blockade of RBD-Omicron binding to ACE2 by immune sera. The dilution ratio of sera was 1:90. Negative: without RBD-Omicron protein; Positive: without sera; PBS: sera from mice immunized with PBS; S1-WT: sera from mice immunized with S1-WT protein; S1-Omicron: sera from mice immunized with S1-Omicron protein. e Flow cytometric analyses of Tfh and GC B cells in spleens. Mice were sacrificed 14 days after the last immunization with PBS, S1-WT, and S1-Omicron recombinant protein vaccines. The spleen lymphocytes were isolated with lymphocyte isolation solution by density gradient centrifugation and stained with cell-surface markers. Tfh cells were identified as CD3+CD19−CD4+CXCR5+PD-1+. GC B cells were CD3−CD19+CD95+GL7+. f, g T-cell recall responses in spleen lymphocytes separated from S1-WT or S1-Omicron immunized mice. NIH mice immunized with PBS, candidate S1-WT or S1-Omicron vaccines were euthanized 14 days after the last immunization. Spleen lymphocytes were isolated and stimulated with recombinant S1-WT or S1-Omicron proteins for 72 h. S1-WT- or S1-Omicron-reactive memory CD4 or CD8 T cells were measured by gating on CD45R−MHC-II−CD44+, as well as CD4+ or CD8+. h Cytokines IFN-γ and IL-4 produced by isolated spleen lymphocytes in culture supernatant were estimated by ELISA after stimulation with 10 μg/ml recombinant S1-WT or S1-Omicron proteins for 72 h. i An overview of the SARS-CoV-2 spike-trimer structure. The N-terminal domain (NTD), receptor-binding domain (RBD), and subdomain (SD-1 and -2) in one protomer are colored green, cyan, and pale yellow, respectively, and marked. j–l A surface view of the known antigenic-sites/epitopes in spike. In each case, one representative neutralizing antibody targeting the site is selected to depict the epitopes (highlighted in red). The mutations identified in the Omicron variant (based on covariant 21 K at address http://covariants.org) are labeled and further marked with arrows. j The supersite in NTD (based on antibody S2M28). k An antigenic site identified in SD (based on S3H3). l The antigenic-sites RBS-A (based on CB6), RBS-B (based on CV07-250), RBS-C (based on CV07-270), RBS-D (based on REGN10987), S309 (based on C135), and CR3022 (based on H014) in RBD. m A schematic view summarizing the Omicron spike-mutations that are in the close proximity to or are directly located within the above-listed antigenic-sites/epitopes. Data are mean ± S.E.M. p-values were determined by unpaired Student’s t-tests (n = 5 in each group). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. ns: not significant