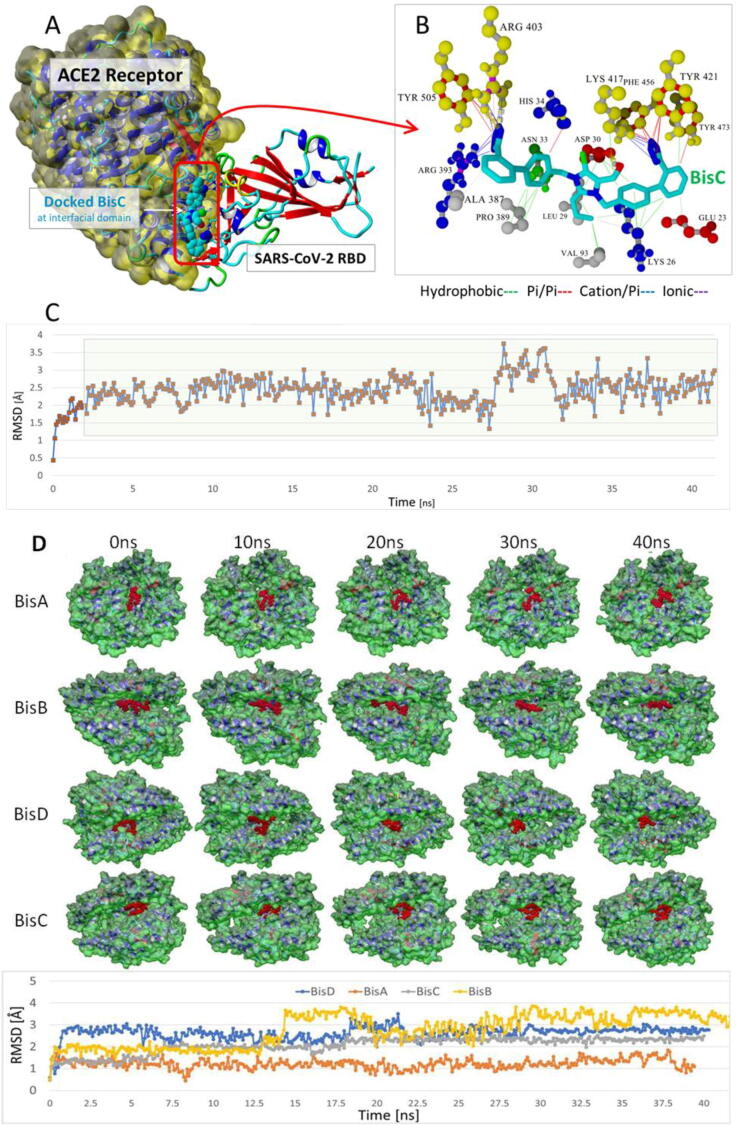
Fig. 6.
(A) Docked chlorinated bisartan BisC to the interfacial region between the ACE2 receptor (Van der Waals surface; yellow) and the SARS-CoV-2/RBD (PDB 6LZG). This pose resulted from the global docking of BisC to PDB entry 6LZG using AutoDock VINA. The docking domain comprised cuboidal cells with non-periodic (wall) boundaries 8 Å from any target atom. (B) The BisC binding motif primarily involved pi/pi (red lines), pi/cation (blue lines), and hydrogen bonding (thick dashed lines) interactions with the SARS-CoV-2 RBD residues Tyr505, Arg403, Phe456, and Tyr421 (yellow spheres). The binding of BisC to the ACE2 interfacial region was mainly dominated by hydrophobic interactions (green lines) to Pro389, Leu29, Val93, Lys26, and Asp30; and secondarily by pi/cation interactions (blue lines) to Arg393. (C) The bound BisC molecule was moderately stable and remained bound inside the interfacial zone (ave. RMSD = 2.46 Å) in MD simulations run out to at least 41 ns (NPT ensemble, 0.9% saline, 311°K). The green shading indicates the time period over which the RMSD values were calculated. (D) Upper panel: Frame captures from MD simulations of bisartan-A, B, C, D/ACE2 complexes. Bound bisartans are indicated by red spheres. MD simulations were run with periodic boundaries for approximately 40 ns at 311oK in physiological saline (water and NaCl ions are hidden for clarity). All bisartans remained stable inside the zinc pocket of ACE2, although Bis B, C, and D exhibited greater motion (RMSD[ave] = 3.2 Å, 2.1 Å and 2.6 Å respectively) compared to BisA (RMSD[ave] = 1.22 Å). ACE2 molecular surfaces are shown. Lower panel: RMSD values as a function of MD simulation time. The stability of the complexes is retained for at least 40 ns.