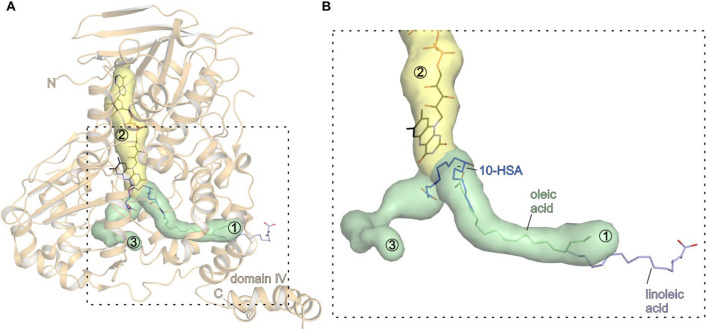
Fig. 5.
Substrate, product tunnel and FAD-binding site in OhySa. A OhySa in light orange cartoon representation and the predicted substrate, product and FAD cavities in surface representation in yellow and green. The substrate/product channel from the exterior of the protein towards the FAD is shown in green and numbered “3”. The cavity with bound FAD is depicted in yellow and numbered with “2”. A side channel in vicinity of the FAD cavity is labelled with “3”. The tunnels were calculated with Caver 3.0 [62]. FAD is shown in black stick representation. Oleic acid (PDB ID: 7kaz; [47]) is shown in light green stick representation; 10-HSA bound to OhySa (PDB ID: 7kaz; [47]) is shown in dark blue stick representation as well as linoleic acid bound to OhyLa (PDB ID: 4ia6; [26]) is shown in light violet stick representation; Dashed box displays magnification area as shown in B. B Magnification of dashed box in A with the protein omitted. The ligands linoleic acid, oleic acid and polyethylene glycol are depicted as in A. The ligands guide the substrate channel from the distal end of the channel to the proximal catalytic cleft close to the FAD