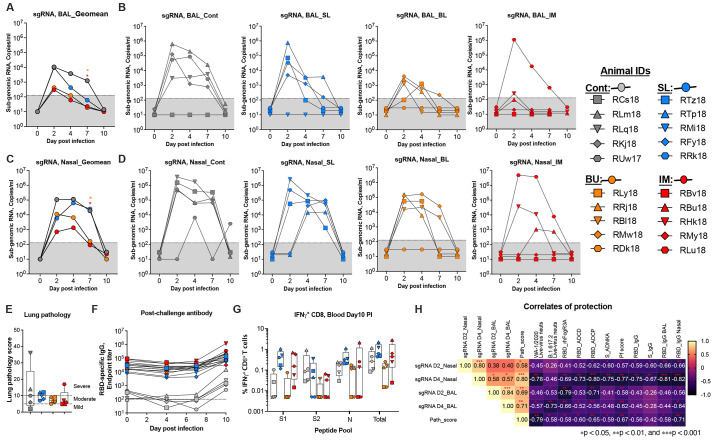
Fig. 6.
Efficacy of MVA/SdFCS-N vaccination against upper and lower respiratory airway SARS-CoV-2 delta (B.1.617.2) viral replication. (A–E) Rhesus macaques vaccinated as per Fig. 3A were challenged with SARS-CoV-2 delta (B.1.617.2) virus via intranasal (IN) and intratracheal (IT) route at week 8, and compared to MVA/Empty immunized controls. Quantified subgenomic RNA (sgRNA) of SARS-CoV-2 in bronchoalveolar lavage (BAL) fluid (A-B), and nasopharyngeal (NS) swab (C-D) collected on days 2, 4, 7 and 10 after challenge and presented as copies/ml. Geomean subgenomic viral loads (A and C) and individual animals subgenomic viral loads (B and D). (E) Lung pathology scores at day 10 post-infection. Hematoxylin and Eosin-stained lung sections were used to analyze tissue structure and cell infiltration. (F) RBD (WA-1/2020)-specific serum IgG responses in serum following challenge. Data represent one independent experiment. Each sample was tested in duplicates. (G) S1, S2 and N-specific CD8+ T cells in blood at day 10 post-infection after re-stimulation with a peptide pool. S1, S1 region of spike residues 1-685; S2, S2 region of spike residues 686-1273; N, nucleocapsid; Total, total response (S1 + S2 + N). (H) Correlation matrix analysis between sgRNA loads in nasopharyngeal (NS) swabs and bronchoalveolar lavage (BAL) fluids, versus 2 weeks post-boost (peak) timepoints RBD (WA-1/2020)-specific binding isotypes, and FcγR binding antibody-dependent functions (neutralization, ADCD, ADCP, ADNP, ADNKA, and polyfunctionality). The color refers to r value scale (−1 to 1) shown on the right. The number in each cell indicates the actual r value and the stars represent p-values. Whisker plots show the maximum and minimum values. Dotted lines in A to D, and F indicate the limits of detection for the assay. Data are mean ± SEM. A two-sided Mann–Whitney U-test was used to compare between groups, *p < 0.05 and **p < 0.01.