Figure 1. Generation of a SB candidate gene list of 136 potential candidate genes in three categories from 4 sources.
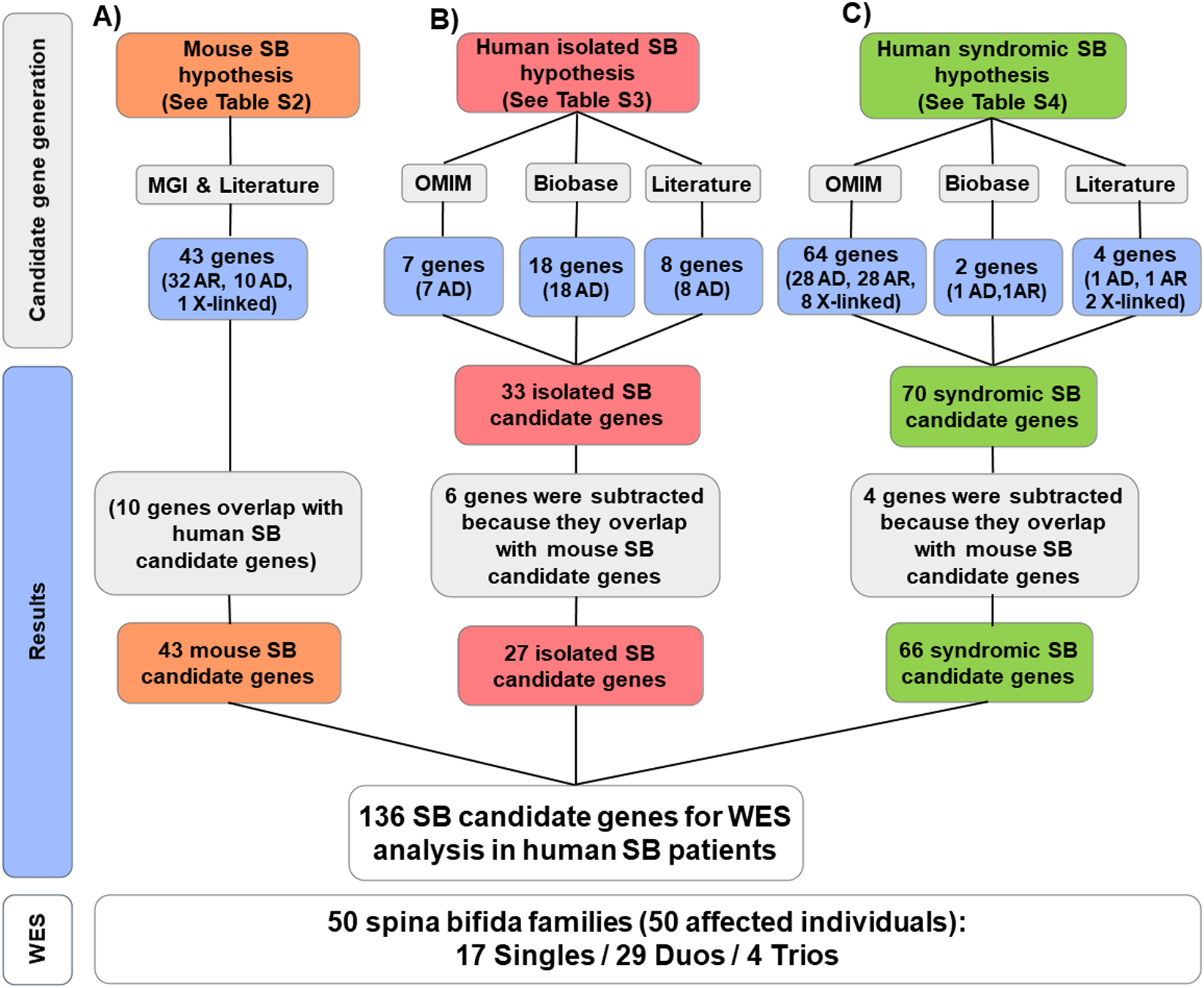
The following databases, MGI (Mouse Genome Informatics; http://www.informatics.jax.org/), Literature (See Methods), OMIM (Online Mendelian Inheritance in Man; https://www.omim.org/), and Biobase-HGMD (Human Gene Mutation Database; https://portal.biobase-international.com/hgmd), were searched to generate a set of four lists of SB candidate genes from four categories of candidate hypotheses: A) 43 candidate genes were derived from mouse models of SB (Table S2); B) 27 candidate genes were derived from existing evidence to potentially cause human non-syndromic SB (Table S3); and C) 66 candidate genes were derived from known causes of human clinical syndromes with facultative SB features (Table S4). Note that we consider candidate status derived from mouse SB as the strongest hypothesis, and since 10 genes in this category overlapped with category B) and C), 6 genes were subtracted from B (non-syndromic human SB candidate genes, red color), and 4) genes were subtracted from C (mouse SB candidate genes, green color). A total of 50 affected individuals with spina bifida from 50 unrelated families were subjected to WES, and their WES data were analyzed for variants in these 136 potential candidate genes (lower panel).
AD, autosomal dominant; AR, autosomal recessive; SB, Spina bifida.
