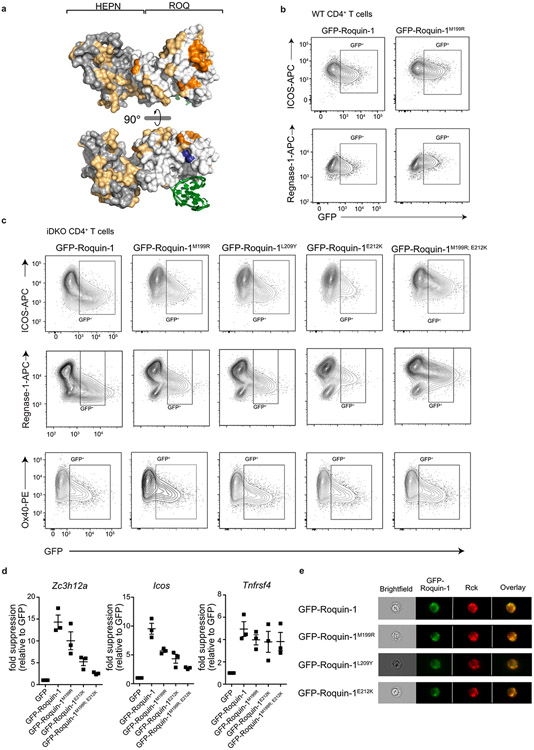
Extended Data Fig. 6 ∣. Functional validation of point mutations in Roquin-1 that reduce cooperation with Regnase-1.
(a) Structure of the Roquin-1 HEPNN/ROQ/HEPNC domain with a bound RNA stem-loop marked in green. All amino acids tested are colored, essential amino acids for Roquin-1 and Regnase-1 interaction in the ROQ domain are marked in orange, non-essential ones in yellow and amino acid M199 in blue. WT (b) or iDKO (c) CD4+ T cells were retrovirally transduced with GFP, GFP-Roquin-1 or the indicated GFP-Roquin-1 mutants. Contour plots of flow cytometry measurement of indicated targets in GFP+ cells after 16 h of doxycycline induction of GFP-tagged constructs. (b) Contour plots of histograms shown in Fig. 4f or (c) shown in Fig. 4g. (d) iDKO CD4+ T cells were retrovirally transduced with GFP, GFP-Roquin-1 or the indicated GFP-Roquin-1 mutants and sorted for GFP+ cells 6 h after doxycycline induced expression of the respective constructs. The levels of Zc3h12a, Icos or Tnfrsf4 mRNAs in GFP+ cells were determined via RT-qPCR, normalized to YWHAZ and calculated as fold suppression of the respective construct relative to cells expressing the GFP control construct. Data show mean +/− SEM of n = 3 independent experiments. (e) Representative images of iDKO CD4+ T cells transduced with the indicated GFP-Roquin-1 WT or mutant proteins, stained with anti-Rck (P-body marker) antibody and analyzed via Image Stream. (b, c) Data are representative of n = 3 independent experiments or (e) n = 2 independent experiments.