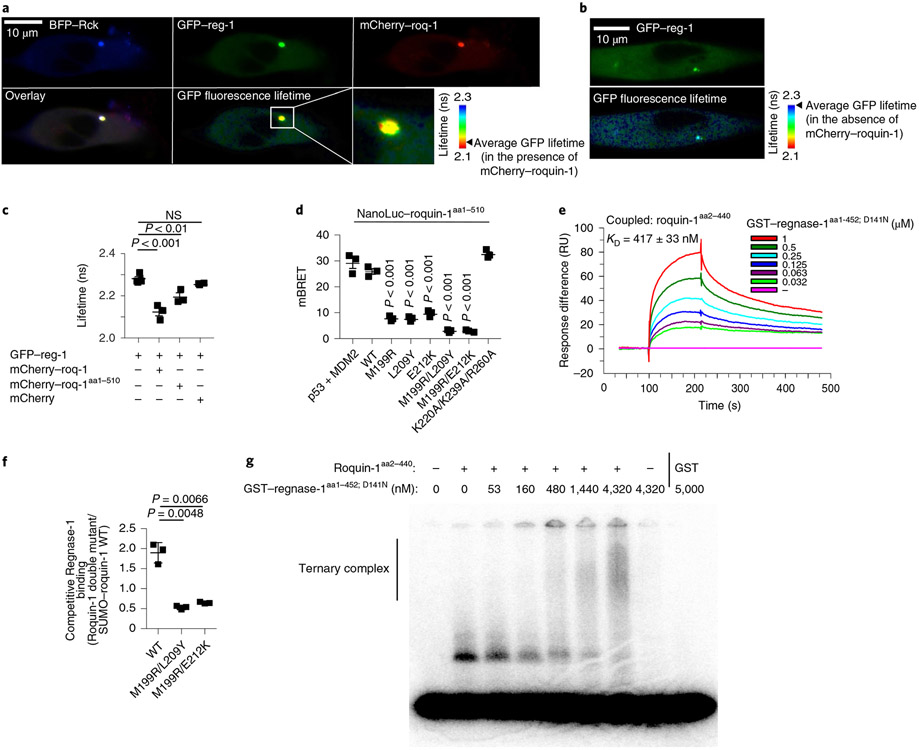
Fig. 5 ∣. Molecular determinants of Roquin-1 interactions with Regnase-1.
a–c, HeLa cells were co-transfected with BFP–Rck, GFP–regnase-1 (GFP–reg-1) together with mCherry–roquin-1 (mCherry–roq-1), mCherry–roquin-1aa1-510 or mCherry (a,c) or with GFP–regnase-1 alone (b). Protein localization (a,b), FRET efficiency (GFP–regnase-1 in combination with mCherry 0.67%, mCherry–roq-1 6.6% or mCherry–roq-1aa1-510 3.91%) and lifetime of GFP fluorescence (c) was analyzed via fluorescence lifetime microscopy. d, HEK293T cells were transfected with HaloTag–regnase-1 in combination with the indicated NanoLuc–roquin-1aa1-510 expression plasmids and NanoBret ratio (mBRET) was calculated after measuring NanoLuc and HaloTag signals via NanoBret assay. e, SPR signals after addition of GST–regnase-1aa1-452;D141N to Biacore-chip-immobilized Roquin-1aa2-440. RU, resonance units. f, Competitive in vitro GST-pulldown experiment using GST–regnase-1D141N and wild-type SUMO–roquin-1aa2-440 in combination with the indicated Roquin-1aa2-440 double mutants (untagged). Quantification of eluted Roquin-1aa2-440 mutant relative to SUMO–roquin-1aa2-440 wild-type protein of SDS–PAGE depicted in Extended Data Fig. 7f. g, EMSA using a Zc3h12a 3′-UTR RNA fragment (nt194–212), Roquin-1aa2-440 (320 nM) in combination with increasing levels of GST–regnase-1aa1-452; D141N. Presumably due to inclusion of unlabeled competitor RNA, recognition of the Zc3h12a mRNA stem loop by Regnase-1 could not be detected. Representative data of n = 3 independent experiments (a,b,e,g). Data are presented as mean ± s.e.m. of n = 3 independent experiments (c,d) or mean ± s.d. of n = 3 independent experiments (f). Statistical significance was calculated using one-way ANOVA with Bonferroni post hoc test (c,d) or Student’s t-tests (f).