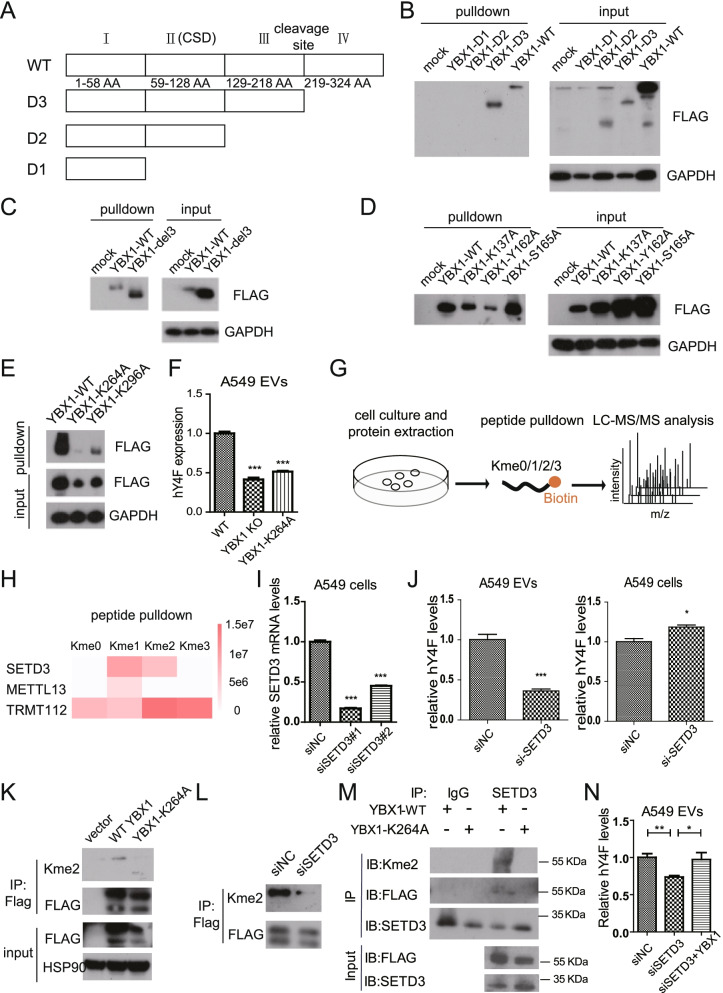
Fig. 6.
SETD3 regulates the sorting of hY4F into EVs through K264 methylation on YBX1. A Schematic representation of the functional domains of YBX1. B-E Biotin-labelled hY4F RNA pulldown analysis with cell lysate from A549 cells overexpressing FLAG-tagged truncated or mutated YBX1. F Quantitative PCR data of hY4F in EVs from YBX1-KO A549 cells transfected with wild type (WT) or K264A mutant YBX1. G Schematic representation of the pulldown assay using biotin-labelled unmodified or mono/di/tri-methyl-lysine modified YBX1-K264 peptide. H The quantitative mass spectrometry results of peptide pulldown assay shown as heatmap. I Verification of siRNA-mediated knockdown of SETD3 in A549 cells by qPCR. J The effect of siRNA-mediated SETD3 knockdown on hY4F level in EVs and cell lysates from A549 cells transfected with SETD3 siRNA. K Methylation of lysine K264 identified by di-methyl-lysine (Kme2) antibody. L The effect of SETD3 knockdown on YBX1 methylation level was assessed by western blot. M Analysis of YBX1 methylation by SETD3 in vitro using endogenous immunoprecipitation assay using SETD3 antibody in the presence of wild-type or K264A mutant. N QPCR analysis of EVs-hY4F levels in A549 co-transfected with YBX1 expression plasmid and siRNA targeting SETD3. Data from three independent experiments are shown as the mean ± SD (error bars). *P < 0.05, **P < 0.01, ***P < 0.001 (Student’s t-test)