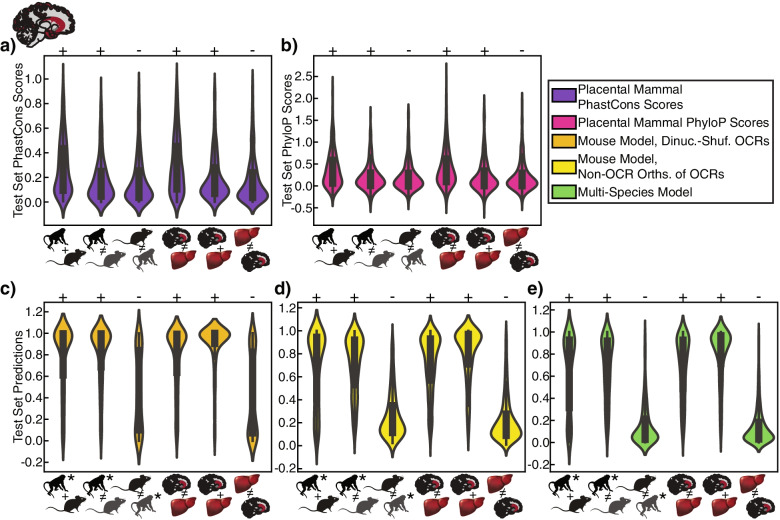
Fig. 2.
Violin Plots for Brain Model Lineage-Specific and Tissue-Specific OCR Accuracy Evaluation in Macaque. Comparison of a PhastCons [13] and b PhyloP [14] scores to c-e three different machine learning models’ predictions for brain OCRs with conserved open chromatin across mouse and macaque, macaque brain OCRs whose mouse orthologs are closed in brain, macaque brain non-OCRs whose mouse orthologs are open in brain, macaque brain OCRs that are closed in liver, macaque brain OCRs that are open in liver (centered on brain peak summits), and macaque liver OCRs that are closed in brain. + ’s indicate that values should be large, and -‘s indicate that values should be small. Conservation scores were generated from the mm10-based placental mammals alignment [12, 73] and averaged over 500 bp centered on peak summits, where mouse peak summits were used for OCRs conserved between mouse and macaque and for OCRs in mouse whose macaque orthologs are closed, and mouse orthologs of macaque peak summits were used for other evaluations. All machine learning model predictions were made using macaque sequences. The macaque sequences for OCRs conserved between mouse and macaque and for OCRs in mouse whose macaque orthologs are closed were centered on macaque orthologs of mouse peak summits, and macaque peak summits were used for other evaluations. Note that the models in c and d were trained on only mouse sequences, demonstrating their performance in a species not used in training. Animal silhouettes were obtained from PhyloPic [65]. *’s indicate the species from which sequences were obtained for making predictions. Dinuc.-shuf. stands for dinucleotide-shuffled, and orths. stands for orthologs