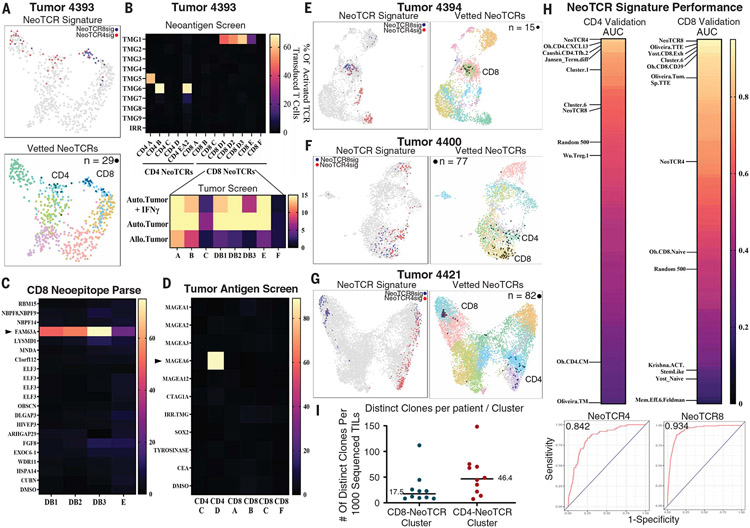
Fig. 3. NeoTCR signatures enable prospective identification of neoantigen-, tumor-associated antigen-, and tumor-reactive CD4 and CD8 TCRs from newly resected tumors.
(A) (Top) Transcriptomic map of 630 TILs sequenced from tumor 4393 (colon). Cells that score in the 95th percentile of NeoTCR4 and NeoTCR8 signatures are highlighted. (Bottom) Experimentally verified CD4+ and CD8+ NeoTCR-expressing cells (n = 29) backprojected onto a transcriptomic map (black). (B) (Top) Screening of eight predicted candidate CD8+ TCRs and six candidate CD4+ TCRs against peptide pools and TMGs that represent 156 tumor mutations identified four TMG- and peptide pool–reactive CD8+ TCRs and three TMG- and peptide pool–reactive CD4+ TCRs. TMGs are shown for clarity. (Bottom) Screening of CD8+ TCRs against corresponding autologous tumors demonstrated selective reactivity toward autologous (Auto.) tumors relative to allogeneic (Allo.) tumors by 7 of 8 TCRs. The right axis, which shows the percentage of activated TCR transduced cells, refers to cells expressing 4–1BB as determined by flow cytometry. (C) Reactivity deconvolution data of the TMG-reactive CD8+ TCRs against constituent peptides of their reactive TMGs for newly identified TCRs from tumor 4393. CD8+ TCRs reactive to TMG1 (TCRs DB1, DB2, DB3, and E) demonstrated reactivity to FAM63Amut (p.D460N). (D) Screening of neoantigen–nonreactive TCRs from tumor 4393 against autologous dendritic cells expressing TAA RNA or pulsed with TAA peptide pools identified tumor 4393 CD4 TCR D as reactive toward MAGEA6. (E) (Left) Transcriptomic map of 2972 TILs sequenced from tumor 4394 (colon). Cells that score in the 95th percentile of NeoTCR4 and NeoTCR8 signatures are highlighted. (Right) Experimentally verified CD8+ NeoTCR-expressing cells (n = 15) backprojected onto a transcriptomic map. (F) (Left) Transcriptomic map of 2559 TILs sequenced from tumor 4400 (colon). Cells that score in the 95th percentile of NeoTCR4 and NeoTCR8 signatures are highlighted. (Right) Experimentally verified CD4+ and CD8+ NeoTCR-expressing cells (n = 77) backprojected onto a transcriptomic map. (G) (Left) Transcriptomic map of 10,049 TILs sequenced from tumor 4421 (colon). Cells that score in the 98th percentile of NeoTCR4 and NeoTCR8 signatures are highlighted. (Right) Experimentally verified CD4+ and CD8+ NeoTCR-expressing cells (n = 82) backprojected onto a transcriptomic map. (H) AUC scores showing the sensitivity and specificity of NeoTCR4, NeoTCR8, and published signatures in calling verified tumor- and neoantigen-reactive TCRs from prospective tumor samples. scGSEA was performed on T cells from samples 4393, 4394, 4400, and 4421, and ROC curves were generated to compare signature sensitivity and specificity. AUC values of all signatures were ranked by their ability to call CD4+ NeoTCRs (top left) and CD8+ NeoTCRs (top right). Selected signatures of interest are highlighted. (Bottom) ROC curves of highest-scoring signatures for CD4 (NeoTCR4; left) and CD8 (NeoTCR8; right) NeoTCRs. (I) Dot plots showing numbers of clones present in the C1 NeoTCR4 and C6 NeoTCR8 states per tumor from Fig. 1A, normalized to 1000 sequenced T cells for each tumor. Bars denote median values.