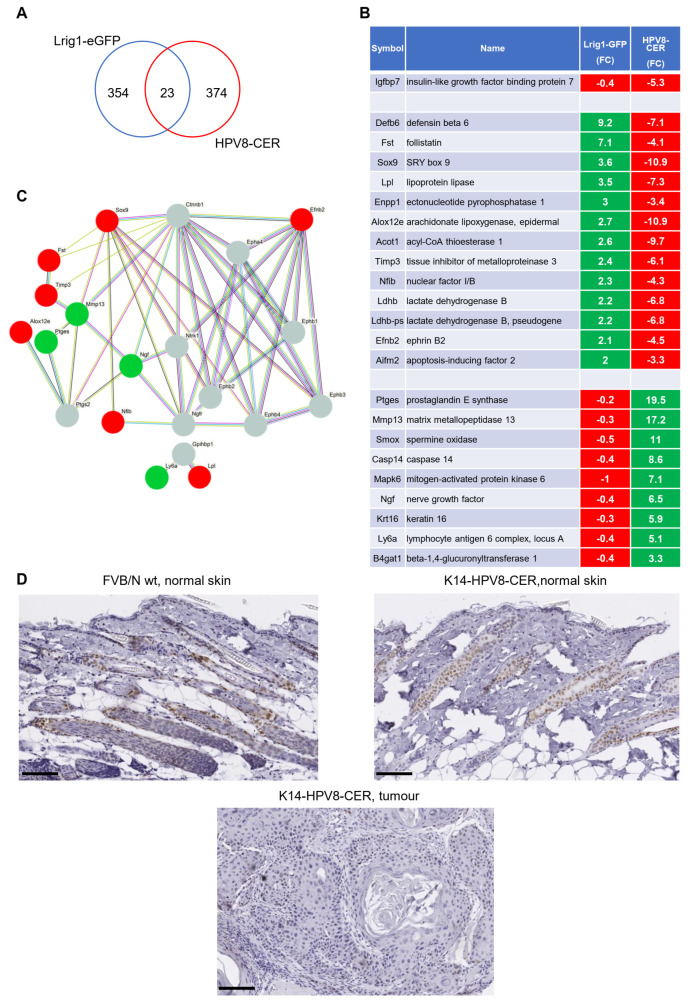
Figure 3.
Comparison of DEGs differentially expressed in K14-HPV8-CER and Lrig1-EGFP-ires-CreERT2 mice. (A) Venn diagram showing the overlap of DEGs found in skin tumours of K14-HPV8-CER and the skin of Lrig1-EGFP-ires-CreERT2 mice. (B) Heat map of 23 genes identified by Venn analysis (green: upregulated genes, red: downregulated genes). (C) The protein–protein interaction (PPI) network plot for the 23 genes using STRING database (code colour: green, upregulated; red, downregulated; grey, database predicted interactors). Line colour indicates the type of interaction evidence e.g., known interaction, or predicted interaction. (D) Representative immunohistochemical staining for SOX9 in FVB/N wild-type, K14-HPV8-CER non-lesional skin as well as K14-HPV8-CER skin tumour. Staining revealed strong expression in FVB/N wild-type and non-lesional K14-HPV8-CER skin. SOX9 was found to be almost completely absent in K14-HPV8-CER tumours (scale bar: 100 μm).