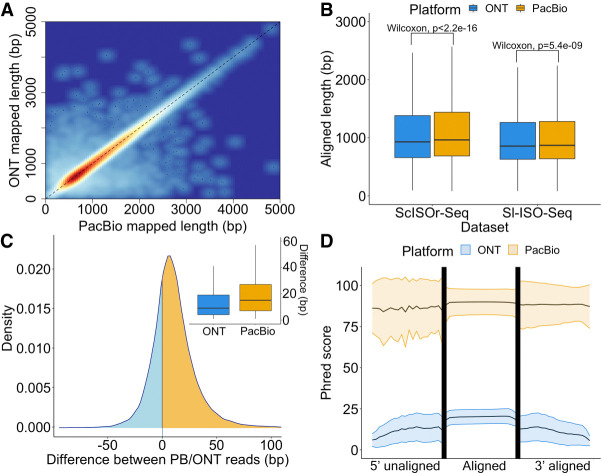
Figure 2.
Alignment characteristics of RT read pairs. (A) Heatscatter plot showing aligned lengths of respective PacBio read (x-axis) and ONT read (y-axis) from the RT read pair after mapping to the genome using minimap2 (Sl-ISO-Seq data set, Spearman's Rho: 0.96, p < 2.2×10−16). (B) Comparison between aligned lengths of PacBio and ONT reads for both data sets after mapping to the genome using minimap2. (C) Density plot showing the difference between aligned lengths of PacBio read and ONT read from the RT read pair after mapping to the genome using minimap2 (Sl-ISO-Seq data set) and box plot showing distribution of the differences. In the density plot, cases when PacBio alignment is longer correspond to the yellow area under the curve; the opposite is represented by the blue area. (D) Mean Phred score distribution along the read for aligned (middle) and unaligned (left and right) parts of PacBio (yellow) and ONT (blue) reads based on a (reference-free) pairwise Smith–Waterman alignment of the PacBio and ONT reads from the RT read pair (Sl-ISO-Seq data set). Lower and upper bounds represent the standard deviation of the Phred score distribution.