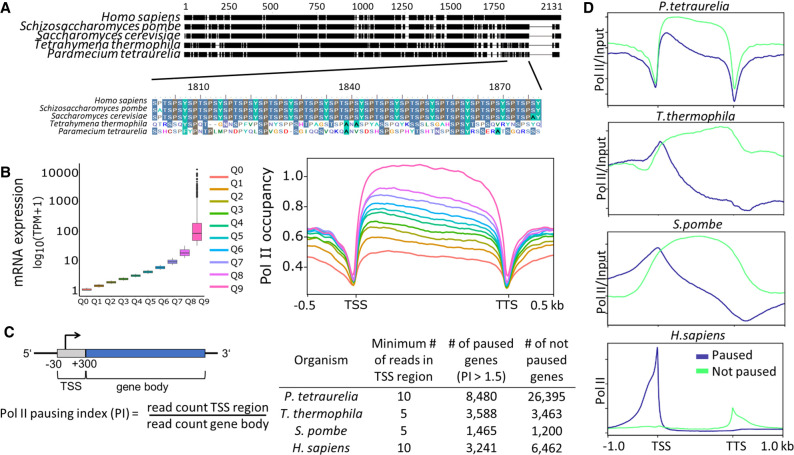
Figure 6.
Analysis of RNA polymerase II pausing. (A) Multiple sequence alignment of the RNA polymerase II enzyme's RPB1 subunit in different organisms is shown. The C-terminal end of RPB1 is zoomed in to show the difference in conserved regions of some ciliates to other organisms. For details, see Supplemental Methods. (B, left) Box plots of gene expression (y-axis; log10 TPM) split in 10 quantiles are shown; higher quantiles mean higher expression. (Right) Pol II enrichment (y-axis) profiles of genes in respective quantiles are shown. Distance shown on the x-axis is scaled; that is, all genes (TSS–TTS) are either stretched or shrunken to a length of 1500 bp. A 500-bp window upstream of and downstream from the gene loci is included. Enrichment profiles were plotted using deepTools2. (C) A graphical representation of the regions included in polymerase pausing index (PI) calculation is shown. We categorized a gene as paused if the PI ≥ 1.5. The table summarizes numbers of paused/not paused genes for selected organisms (Supplemental Table S1 contains details on Pol II data sets). (D) Same as the Pol II enrichment profiles in B, but genes are split based on the status of Pol II pausing.