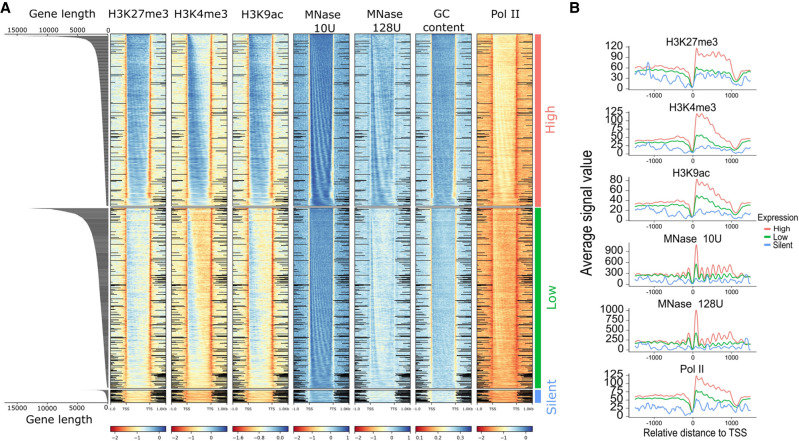
Figure 7.
Distribution of epigenetic marks. (A) Distribution of epigenetic marks in different transcriptomic groups. Heatmaps show the input normalized enrichment values for different epigenetic marks. Genes (rows) are split into three categories based on gene expression—high (TPM > 2), low (0 < TPM < 2), and silent (TPM = 0)—and are sorted by decreasing order of gene length in each, which is visualized by the length distribution graph on the left. Distance shown on the x-axis is scaled; that is, all genes (TSS–TTS) are either stretched or shrunken to a length of 1500 bp, adding 1000 bp upstream of and downstream from the gene. Heatmaps were plotted using deepTools2; black lines in intergenic regions reflect missing data at this position. (B) Distribution of epigenetic marks for a subset of 4000 genes with discrete length of ∼1.2 kb. Plots show the signal in the upstream intergenic region, the TSS and the TTS of genes belonging to the similar expression categories as in B.