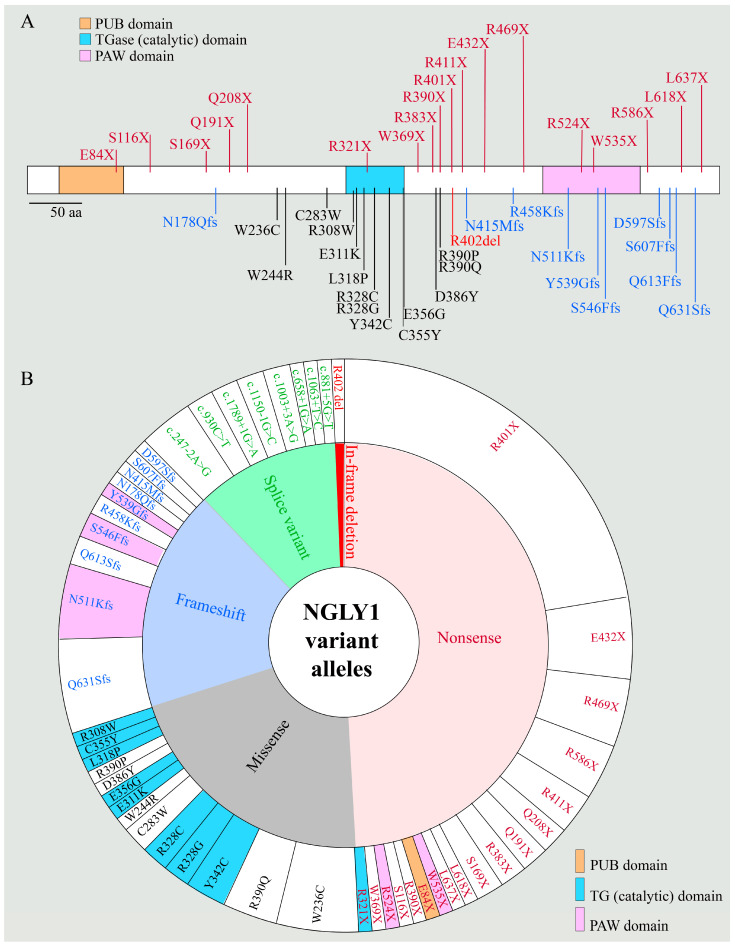
Figure 2.
(A) Schematic representation of the reported missense, nonsense, frameshift, and in-frame deletion variants in NGLY1. Nonsense variants are in maroon font above the NGLY1 schematic. Missense (black font), frameshift (blue font), and in-frame deletion (red font) are shown below NGLY1. Protein structure is based on SMART (simple molecular architecture research tool; http://smart.embl-heidelberg.de accessed on 21 March 2022). (B) Ring chart diagram showing the reported variants, including splice site variants (green font) with their relative occurrence. Note that R401X is the most common variant (~21%). aa, amino acid; fs, frameshift; PAW, a domain in PNGases and other worm proteins; PUB, a domain in PNGase/UBA or UBX-containing proteins; TG, transglutaminase-like.