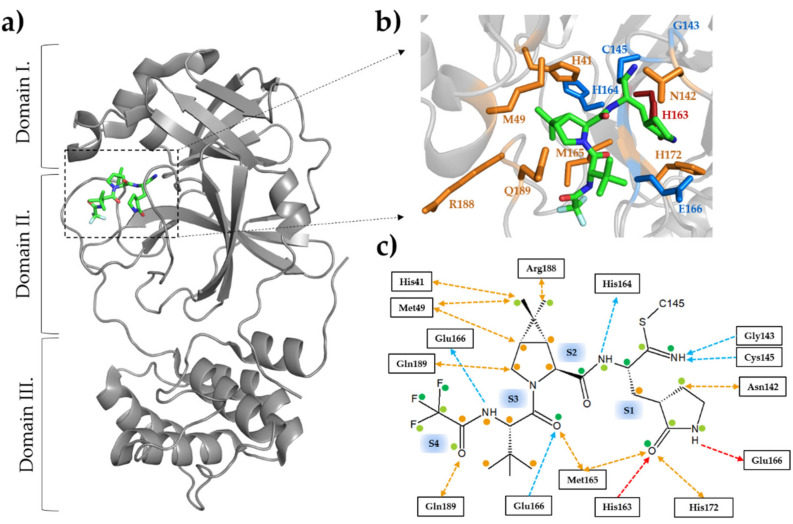
Figure 1.
Structure of SARS-CoV-2 Mpro with nirmatrelvir. (a) The domain organization is represented based on the crystal structure of the enzyme complexed with nirmatrelvir (7RFS.pdb). The inhibitor is represented as sticks, the carbon atoms are green, the oxygen atoms are red, and the nitrogen atoms are blue. (b) Enlarged view of the active site showing the most relevant enzyme-inhibitor interactions. The active-site residues forming at least one main-chain- and side-chain-mediated hydrogen bond are colored by blue and red, respectively. The residues forming apolar interactions are shown in orange. (c) Most relevant inhibitor-binding interactions at the active site. The backbone- and side-chain-mediated hydrogen bonds are colored by blue and red, respectively. The direction of the arrow indicates the donated hydrogen atom. Apolar interactions are shown in orange. Colored dots indicate the average of non-bonded contacts for individual atoms. Orange represents 1 contact, light green represent 2–4 contacts, and dark green represent ≥ 5 contacts. The interactions were mapped based on five crystal structures (7vh8, 7si9, 7te0, 7rfs, and 7rfw) using PDBSum [62] and LigPlot+ [63]. See the details in Table 1. An interaction was considered to be relevant if it was present in at least three of the five studied structures.