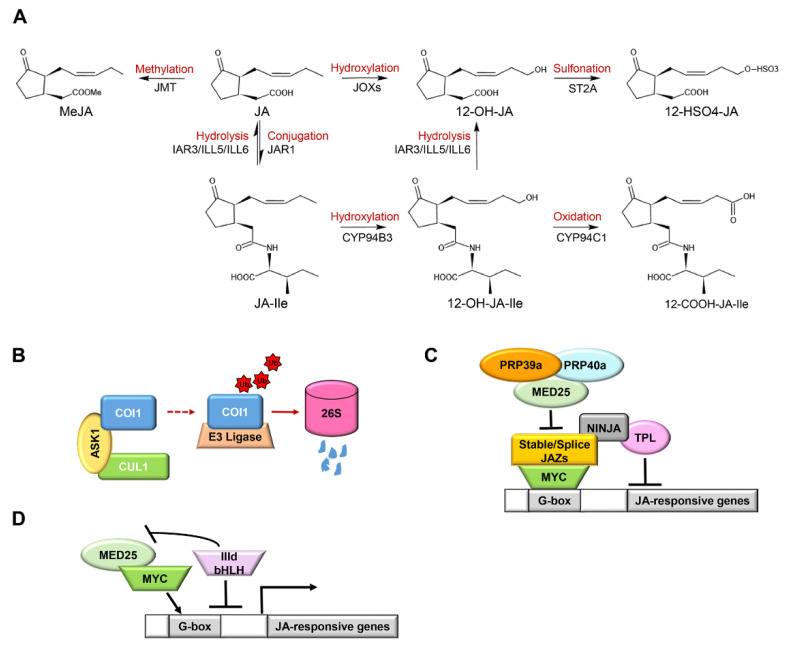
Figure 2.
The terminal regulation mechanism of jasmonic acid (JA) signaling: (A) the dynamic banlance of JAs. Metabolic and catabolic enzymes maintain a dynamic balance between JA and JA-Ile. Methyl jasmonate (MeJA) is formed by JA catalyzed by jasmonic acid carboxyl methyltransferase (JMT). JAR1 transforms JA into JA-Ile. JA-Ile is continuously oxidized twice by CYP94 P450 family enzymes to generate 12-OH-JA-Ile and 12-COOH-JA-Ile in turn. The amide bond of JA-Ile/12-OH-JA-Ile is broken by amide hydrolases (IAR3/ILL5/ILL6) to generate JA/12-OH-JA, respectively. JA can be directly hydroxylated to 12-OH-JA and further sulfated to form 12-HSO4-JA; (B) COI1 degradation pattern. Jasmonate receptor COI1 forms the SCFCOI1 complex with ASK1 and CUL1. Excessive COI1 protein is recruited and degraded by ubiquitination through the 26S proteasome pathway; (C) the inert and alternative splicing of JAZ-mediated JA attenuation pattern. JA signal suppressor JAZ protein can bind to NINJA protein and directly or indirectly recruit co-inhibitor TPL to inhibit the transcription of MYC2, thereby inhibiting the expression of JA-responsive genes. Inert JAZs (JAZ7, JAZ8, and JAZ13) have a conserved EAR domain that directly interacts with TPL. JAZ10.4, a variable splice of JAZ10, lacks the Jas motif and mediates desensitization to JA. MED25-recruited splicing factors PRP39a and PRP40a promote the correct splicing of JAZ genes and prevent the overproduction of splicing variants, thereby regulating the activation of JA signal; (D) bHLH-like protein-mediated JA attenuation pattern. bHLH IIId (JAM1/2/3, bHLH14) proteins compete with MYC-like TFs to bind the G-box element on the promoter of the JA response gene, inhibit the formation of the MYC2-MED25 complex, and negatively regulate the JA response. Arrows: activations; bar-headed arrows: repressions.