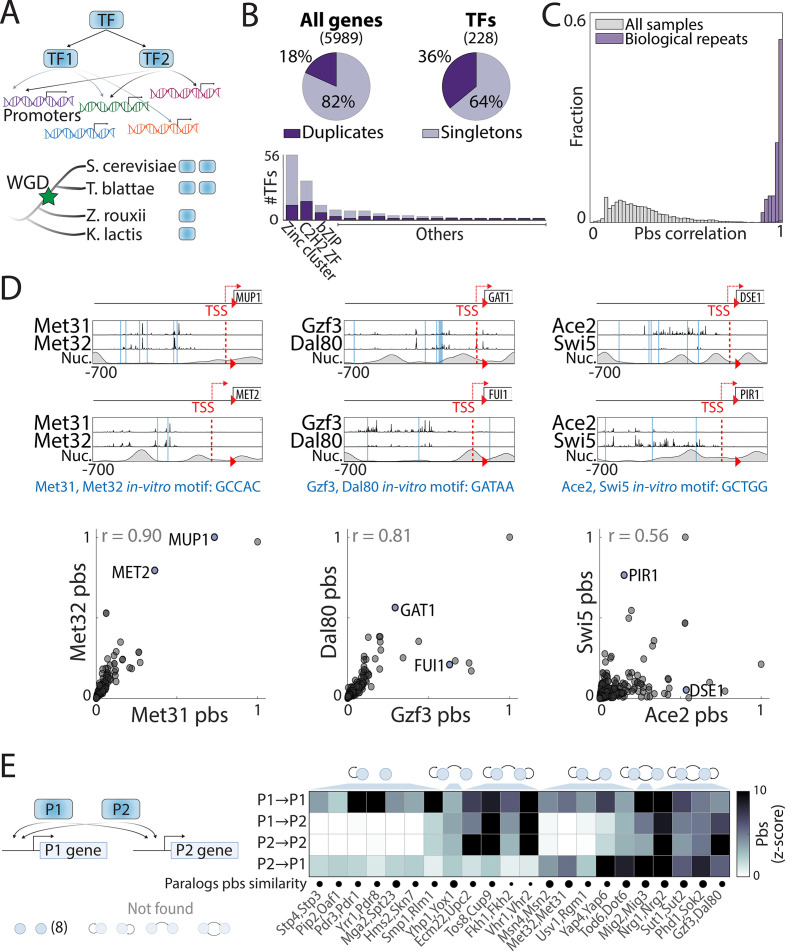
Figure 1. Mapping the promoter-binding preferences of whole-genome duplication (WGD) transcription factors (TFs).
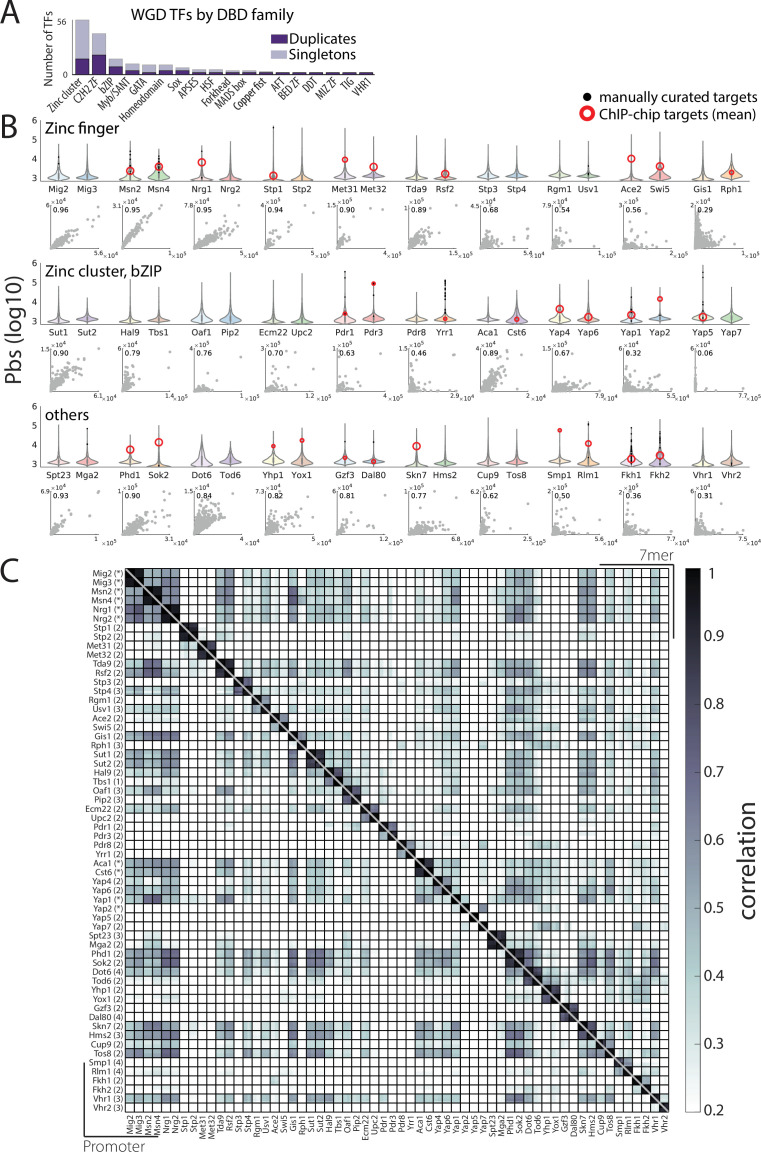
(A–B) WGD shaped the budding yeast transcription network: (A) TF duplicates (paralogs) can diverge to bind different targets. (B) In Saccharomyces cerevisiae, ~35% (Gietz et al., 1995) of all present-day TFs are retained WGD paralogs, belonging to 18 different DNA-binding domain families (see Figure 1—figure supplement 1). (C) TF-binding profiles are reproducible: Shown is the distribution of correlations between different samples (gray) and between biological repeats (purple). Correlations are between promoter-binding signals (pbs). (D) Binding profiles of indicated TF-paralog pairs. Top: Measured binding signal and nucleosome occupancy (Nuc.) on individual promoters (see Materials and methods). Lines indicate transcription start sites (TSS, red dashed) and locations of in vitro motifs (blue). Bottom: Pbs of the indicated TF-paralog pairs (each dot is a promoter, r: Pearson’s correlation). (E) Auto- and cross-promoter binding by TF paralogs: Pbs is shown as z-score. Potentially formed circuits indicated on top. Note that 22/30 pairs are associated with six of nine possible circuits.