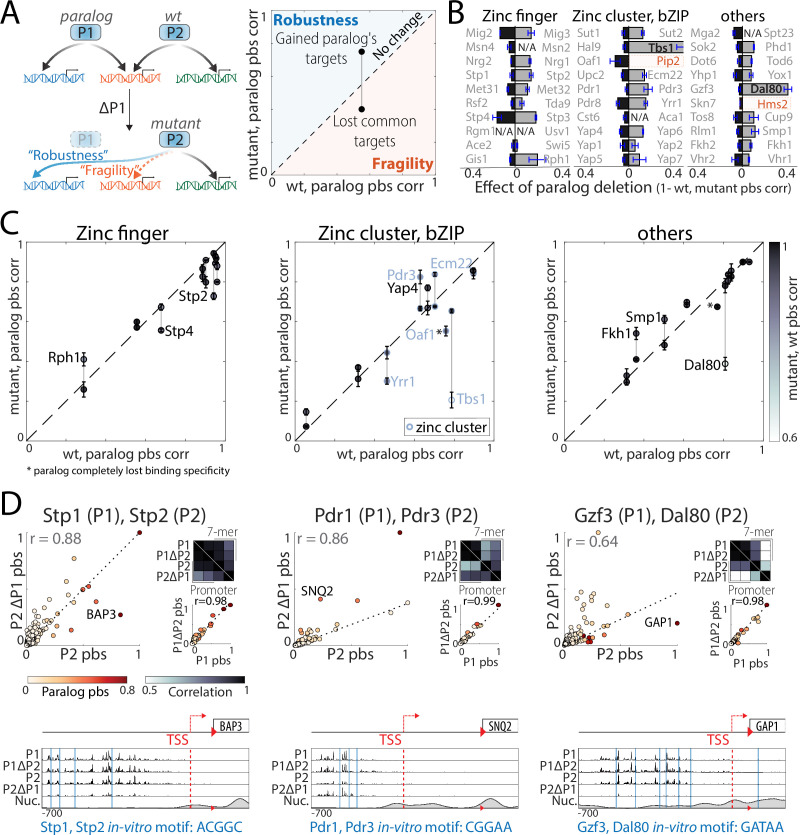
Figure 5. Interactions between transcription factor (TF) paralogs may increase network fragility.
(A) Paralogs’ contribution to mutation robustness or fragility: Following paralog deletion, a TF may gain access to its paralog’s unique sites, potentially compensating for the loss (‘robustness’, blue line: gained paralog target). Alternatively, paralogs may become interdependent and loose common targets after paralog deletion (‘fragility’, dashed orange line: lost common target). At the genome level, these interactions can be summarized by comparing a TF’s binding preferences in wild-type (x-axis) or paralog-deleted (y-axis) backgrounds to those of the paralog. (B) Strong paralog interactions are rare: the effect of paralog deletion on promoter-binding preferences was measured for 55 of 60 TFs in our dataset. Shown is the effect of paralog deletion on binding preferences for each TF. Note that most deletions were of little effect and that large effects were asymmetric. Also indicated are substantial effects (TFs written in black) and TFs that completely lost binding specificity (orange, see Materials and methods; N/A: not profiled). (C–D) Paralog interactions within individual families: (C) robustness/fragility analysis, as in (A) for all tested paralog pairs, divided into families (*: paralog completely lost binding specificity). (D) Shown are individual examples of the depicted correlations (see Figure 5—figure supplement 1 for all tested pairs). Note that Stp2 and Dal80 loose binding to some of their paralog’s targets upon paralog deletion (‘fragility’), whereas Pdr3 gains binding to Pdr1 targets (e.g. SNQ2) upon the latter’s deletion (‘robustness’).