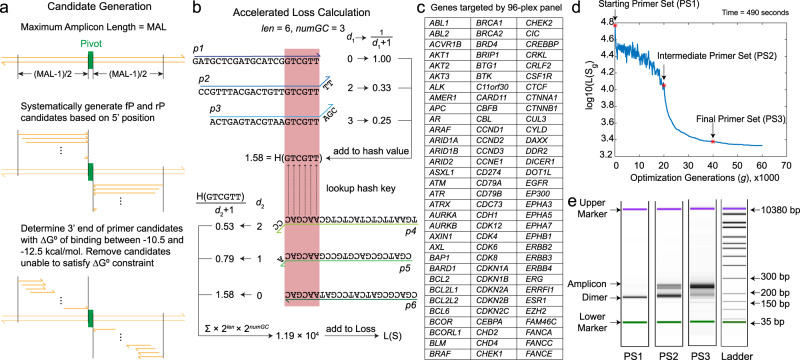
Fig. 2. Implementation and experimental evaluation of a multiplex primer design algorithm based on the SADDLE framework.
a Method for generating candidate primer sequences for a DNA target T. b Implementation of Badness function that can be rapidly evaluated using hash tables. len is the length of the subsequence, numGC is the number of G/C nucleotides in the subsequence. d1 and d2 are the distances of the subsequence to the 3′ ends of primer pa and pb, respectively. p1, p2, p3 are examples of primer pa, and p4, p5, p6 are examples of primer pb. c List of cancer genes selected as target sequences for a 96-plex primer set design. See Supplementary Excel spreadsheet for target selection details. d Loss function of primer sets S(g) across optimization generations g. The Loss function value decreases through the optimization and approaches a local minima after roughly 400 generations. We selected three different primer sets, constructed at generations 0, 200, and 400 for experimental evaluation; these are respectively called PS1, PS2, and PS3 for the remainder of this paper. Computation time for a 96-plex panel design is about 490 s for 60,000 iterations on a conventional laptop. e Capillary electrophoresis (Agilent Bioanalyzer 2100) analysis of amplicon products of PS1, PS2, and PS3. Here, 10 ng of the NA18562 human genomic DNA (Coriell) was used as input, and the median primer concentration was 45 nM in the PCR reaction. Seventeen cycles of PCR were performed using Vent (exo-) DNA polymerase (selected for its improved amplification ability for G/C-rich sequences). To facilitate more in-depth analysis by high-throughput sequencing (NGS), adapters/indexes were ligated to the amplicon products. No size selection was performed, in order to accurately reflect the fraction of primer dimer species following multiplex PCR. The on-target amplicons are expected to have an average length of roughly 250 nt.