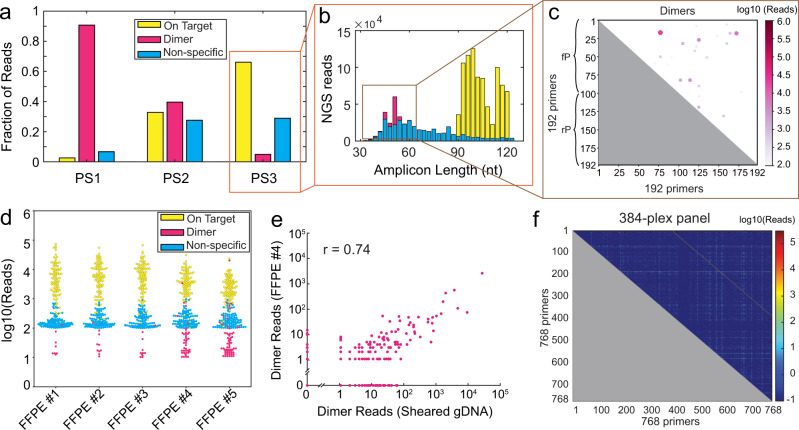
Fig. 3. Experimental NGS results for SADDLE-designed primer sets.
a Distribution of reads observed in NGS library constructed using PS1, PS2, and PS3. On Target reads are defined as those that aligned to the intended amplicons; Dimer reads are defined as those whose insert lengths are smaller than the sum of the two primer lengths; all other reads were classified as Non-specific. The vast majority of Non-specific reads align to other regions of the human genome, via a non-cognate pair of forward and reverse primers. The fraction of NGS reads mapped to Dimers dramatically decreases from PS1 to PS2 to PS3. b Distribution of NGS reads in the three primer set libraries. c Distribution of observed primer dimers, based on aligned reads. Because forward primers (fP) can also form primer dimers with other forward primers, we aligned the first and last 25 nucleotides of each NGS read to the merged set of fP and rPs, with primers 1 through 96 in the diagram showing fPs and primers 97 through 192 showing rPs. For clarity of visualization, the log number of reads of observed primer dimers are displayed via both coloration and circle size. d Performance of the PS3 primer set of formalin-fixed paraffin-embedded (FFPE) tissue samples from deidentified lung cancer patients. Because the NGS libraries for these five samples differed slightly in total reads, here we plotted the distribution of reads normalized to 1 million reads. e The observed primer dimer species and their corresponding NGS reads were relatively similar between cell line genomic DNA and FFPE samples. f Demonstration of a 384-plex primer set designed by SADDLE (768 primers). The main diagonal shows On Target reads. Only about 1% of all reads were primer dimers (Supplementary Section S6).