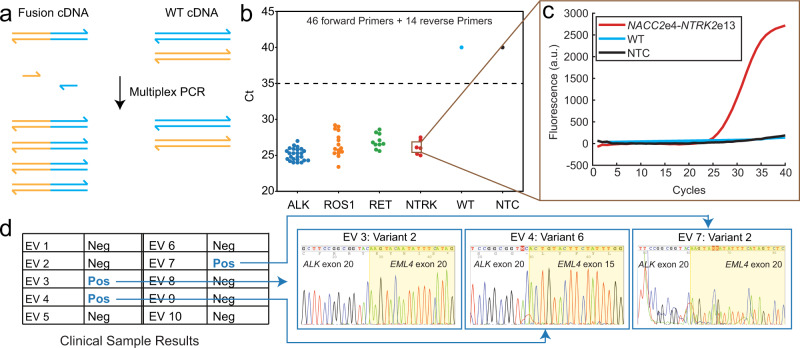
Fig. 5. Highly multiplexed qPCR detection of gene fusions using SADDLE-designed primer sets.
a Complementary DNA (cDNA) prepared through reverse transcription of RNA can have known target sequences at the exon breakpoints. Although it is trivial to design a single-plex qPCR assay to detect a single known fusion, such as BCR-ABL136, we are not aware of any reports of highly multiplexed qPCR assays to simultaneously detect ≥10 different gene fusion cDNA species. For this assay, we designed a 60 primer set (46 forward, 14 reverse) that together can amplify 56 distinct gene fusion types commonly observed in non-small-cell lung cancer37. b Summary of observed qPCR cycle threshold (Ct) values for the 56 reactions, each with 1 of the 56 synthetic fusion DNA species across 6 genes (ALK, ROS1, RET, and NTRK1/NTRK2/NTRK3), each with 1700 copies. WT indicates wildtype commercial cDNA, and NTC indicates no template control. See Supplementary Section S8 for additional details and experimental results, including Sanger sequencing traces of each reaction product. c Example qPCR trace showing detection of the fusion DNA sequence joining NACC2 exon 4 to NTRK2 exon 13. d Clinical sample results on cDNA reverse transcribed from RNA from extracellular vesicles. Samples 3, 4, and 7 tested positive for a gene fusion, and sequence alignment of the Sanger sequencing results (right panels) show the exact identifies of the fusions.