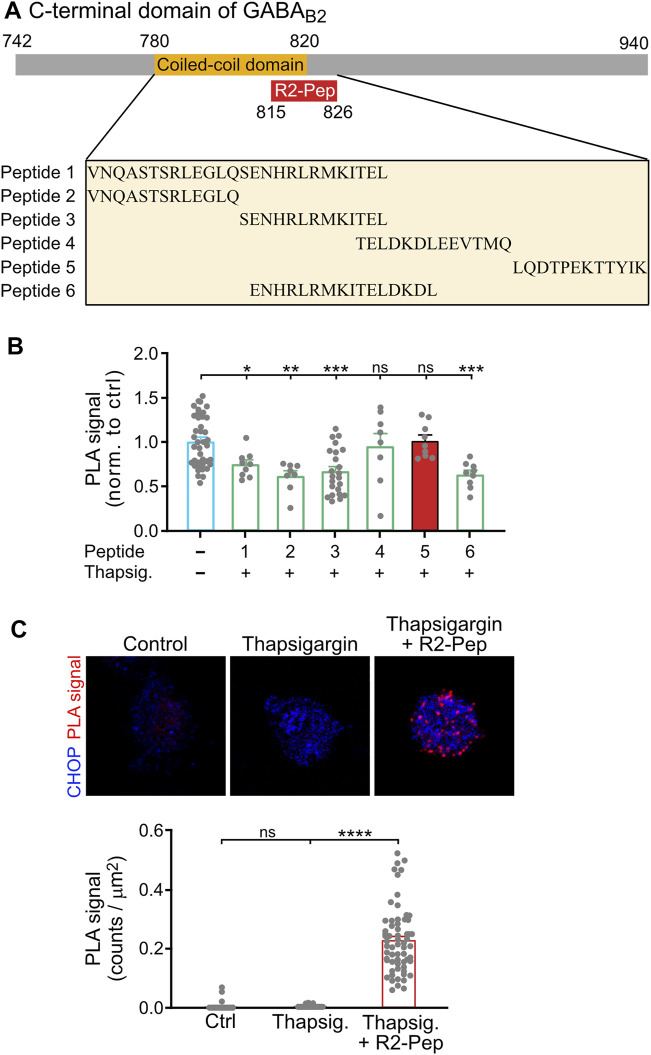
FIGURE 2.
Screening for a peptide (R2-Pep) interfering with the interaction of CHOP with GABAB receptors. (A) Scheme of the C-terminal domain of GABAB2 with the coiled-coil domain (orange box), which contributes to the CHOP-GABAB receptor interaction site and location of the interfering peptide R2-Pep sequence (red box). The box below depicts the GABAB2 peptide sequences used for screening. (B) Peptide 5 reliably prevented stress-induced decrease in GABAB1/GABAB2 heteromers. Cultures were stressed for 2 h with thapsigargin (1 μM) and immediately thereafter treated for 30 min with the peptides indicated in (A). Neurons were then tested for the interaction between GABAB1 and GABAB2 by in situ PLA. The in situ PLA signals of the untreated neurons served as control (-, no peptide). The data represent the mean ± S.E.M. of 8–40 neurons derived from two independent experiments (control, n = 4). ns, p > 0.05; *, p < 0.05; **, p < 0.005; ***; p < 0.0005. Brown-Forsythe/Welch one-way ANOVA followed by Games-Howell’s multiple comparison test. (C) R2-Pep (peptide 5 in B) interacted with CHOP. Cultures were stressed for 2 h with thapsigargin (1 μM) and immediately thereafter treated with R2-Pep for 30 min. Neurons were then tested for the interaction of CHOP with R2-Pep by in situ PLA using antibodies directed against CHOP and FITC (R2-Pep was labeled with FITC at the N-terminus). The data represent the mean ± S.E.M. of 47–62 neurons derived from two independent experiments. ns, p > 0.05; ****; p < 0.0001. Kruskal–Wallis test followed by Dunn’s multiple comparison test.