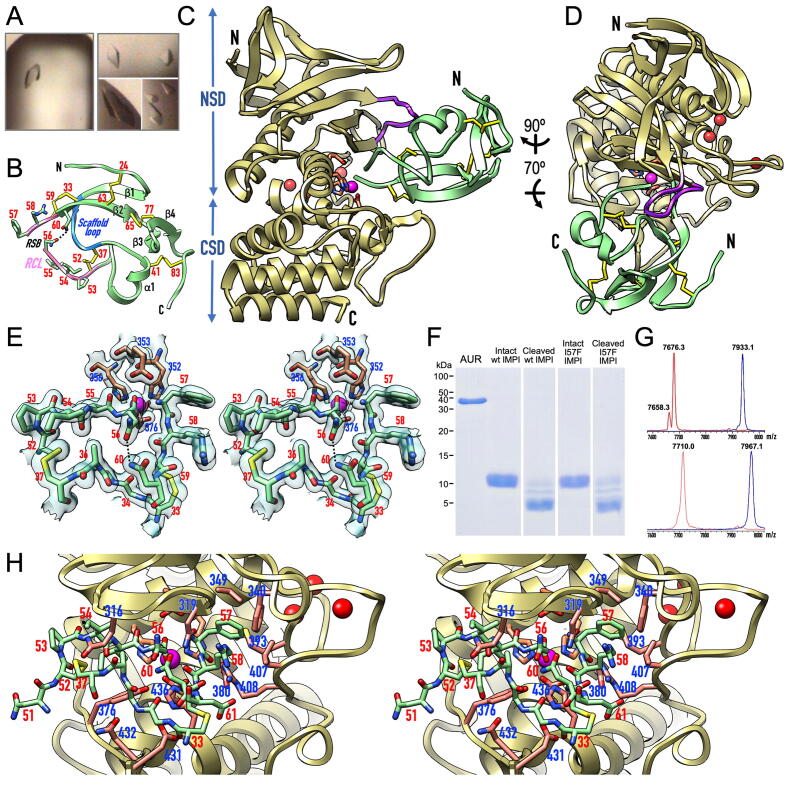
Fig. 3.
Structure of the IMPI–aureolysin complex. (A) Tetragonal protein crystals of the aureolysin–wt-IMPI (left) and aureolysin–I57F-IMPI complexes (right). (B) Ribbon-type plot of I57F-IMPI depicting the four β-strands (β1–β4) and the single helix (α1) of the structure, as well as the five disulfide bridges (with numbered cysteine residues). The scaffold loop is shown in blue, and the reactive-centre loop (RCL) is shown in pink with numbered residues (sticks). The cleaved reactive-site bond (RSB), N-terminus, and C-terminus are labelled. Hydrogen bond N56Oδ1–N60Nδ2 is needed to maintain the position of the P1 residue in place. (C) Ribbon-type plot of the complex between I57F-IMPI (green ribbon, disulfide bonds as yellow sticks) and aureolysin (pale gold ribbon, catalytic zinc and structural calcium cations shown as magenta and red spheres, respectively) viewed along the active-site cleft (vertically rotated 90° counterclockwise away from the traditional “standard orientation” of MPs [53]). The side chains of the zinc-binding MP residues and the general/base acid glutamate are further shown as sticks for reference (carbons in salmon). The N-termini and C-termini are labelled, the characteristic flap is in purple, and the NSD and CSD of the peptidase are indicated. (D) Rotated view of (C). (E) Close-up in cross-eye stereo showing the RCL and scaffold loop of I57F-IMPI (green carbons) and the zinc site of aureolysin (carbons in salmon) superposed with the final 1.60-Å (2mFobs-DFcalc)-type Fourier map as a semi-transparent surface contoured at 1 σ in a similar view to (D). The RSB is cleaved, selective inhibitor and MP residues are numbered in red and blue, respectively. Hydrogen bond N56Oδ1–N60Nδ2 is shown as a dashed line. (F) In vitro proof that binding and inhibition of aureolysin by wt- and I57F-IMPI involves the cleavage of the inhibitor at the RSB (N56–I57) within the RCL as shown by SDS-PAGE analysis of the respective SEC fractions. (G) Mass spectra showing analysis of the cleavage of (top) intact wt-IMPI (blue spectrum; 7933.1 Da) giving rise to the cleaved inhibitor (red spectrum; 7676.3 Da) and (bottom) intact I57F-IMPI (blue spectrum; 7967.1 Da) to yield the cleaved inhibitor (red spectrum; 7710.0 Da). Incubation of both intact species with aureolysin leads to the removal of the N-terminal tag-segment G-M−S (–275 Da) and the addition of a water molecule (+18 Da) due to RSB cleavage. For wt-IMPI, a small fraction of tag-depleted noncleaved inhibitor was detected (7658.3 Da). (H) Close-up in stereo of (D), further rotated 25° downwards and 25° leftwards, giving insight into the interactions between I57F-IMPI (sticks with green carbons, residue numbers in red) and aureolysin (sticks with carbons in salmon, residue numbers in blue). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)