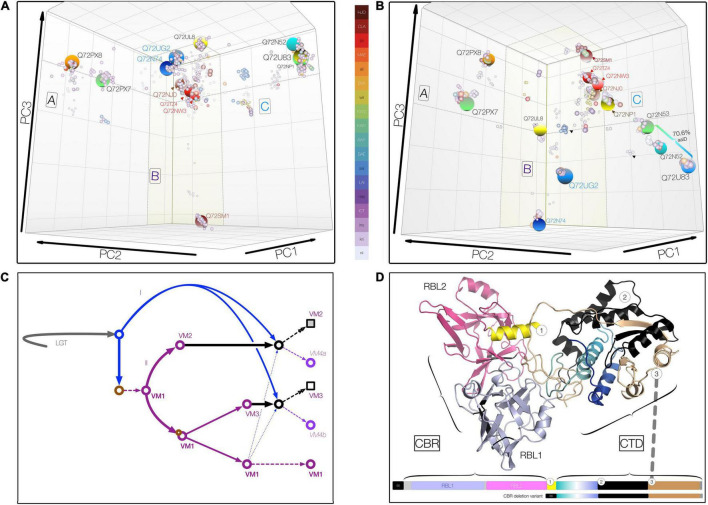
FIGURE 2.
Three-dimensional metric multidimensional scaling (3DMMDS/Galaxy) plots depicting (orthologous) VM protein clusters. Clusters were identified among 940 PF07598 family VM proteins analyzed using bios2mds (Pele et al., 2012) and visualized using principal component analysis in R. In addition to typical PF07598 paralogs, 42 natural deletion mutants lacking both ricin B-like lectin, RBL, subdomains (i.e., containing amino terminal signal sequence and toxin domain only) were included. (A) Carbohydrate-binding region (CBR), containing two unidentical tandem RBL subdomains. (B) Carboxy terminal toxin domain (CTD) encompassing discrete trafficking and DNase subdomains. For both, initial renderings were edited (cosmetic changes only) to aid visualization by enhancing the 3D effect. No coordinates were altered. Clusters containing VM protein variants found in Leptospira interrogans are highlighted (large spheres) and named using the reference L. interrogans serovar Copenhageni strain (PMID 15028702), L1-130 (UniProtKB) protein IDs. Orthologous clusters were grouped into three superclusters comprising VM protein paralogs (A, n = 2; B, n = 7; and C, n = 4) based upon percent identity (PID) (Supplementary Tables 1–4). The color key uses the following convention: for species, L. interrogans (ins), L. kirschneri (kri), L. noguchii (nii), etc.; for serovar, e.g., Canicola (CLA), Lai (LAI), Hardjo (HJO), etc.; and for strains originating from Sri Lanka, e.g., L. interrogans serovar Unknown strain KW1 (KW1), etc. (C) Schematic showing a theoretical evolutionary history of the VM protein family, involving lateral transfer (LGT), gene duplication (purple arrows, II) and erosion (solid black arrows), and recombination (blue arrows = donor acquired via lateral gene transfer, I; broken arrows indicate intragenomic donor from closely related paralog). Circles represent theoretical evolving VM proteins over time; squares represent final evolved form at the current time. (D) Domain organization and junctions of chimeric Leptospira VM proteins resulting from CBR and CTD domain fusions of paralogs belonging to closely related CBR clusters, such as those related to Q72NW3 (e.g., WP.017856587.1) and Q72TZ4 (e.g., QHH71994.1) (∼99.1% PID, Supplementary Tables 1, 4). These natural VM protein variants occur infrequently (∼2%) in L. interrogans and its sister species, L. kirschneri and L. noguchii. Chimeric VM proteins generally share a common junction regardless of the paralogs represented.