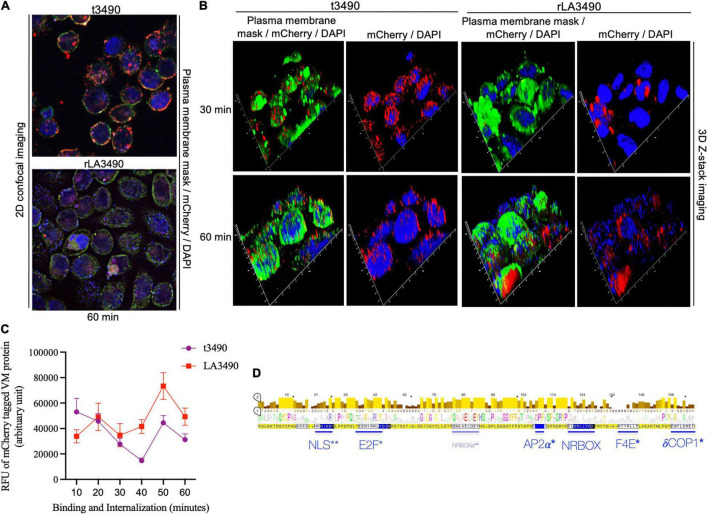
FIGURE 5.
Surface binding and nuclear localization of rLA3490 in HeLa cells. Fluorescent confocal microscopy demonstrating kinetics of binding of mCherry–rLA3490 and mCherry–t3490 fusion proteins binding to HeLa cells. (A) Two-dimensional view, at 60 min, t3490 is visible only on the cell surface (red); rLA3490 is internalized by 60 min (red/pink). (B) Three-dimensional Z-stack and orthogonal images obtained by high-resolution fluorescent confocal microscopy showing internalization of mCherry–rLA3490 fusion from 30 min onward, with nuclear translocation and chromosomal degradation (shown by patchy DAPI staining, lower right) evident within 60 min. t3490 remained on the cell surface at 30 and 60 min. Visualization of treated cells was done after staining with CellMask™ green plasma membrane stain mounting with ProLong™ Gold Antifade Mountant + DAPI nuclear stain. Images were captured using an oil immersion × 100 objective using appropriate filters (blue, DAPI; green, plasma membrane; red, mCherry fusions). (C) Time-dependent interactions of mCherry-tagged rLA3490 and t3490 proteins with HeLa cells (surface binding plus internalization). Fluorescent confocal microscopy (using ImageJ version 1.53 software) was used to quantify recombinant fusion proteins with HeLa cell monolayers. Monolayers were exposed to 45 nM recombinant fusion proteins or controls up to 60 min. Fluorescence intensities of mCherry–t3490 and –LA3490 fusion proteins were measured in 10-min intervals from 0 to 60 min. Data were visualized in GraphPrism 8. (D) Eukaryotic trafficking and protein–protein interaction motifs found in Leptospira VM proteins. Consensus amino acids in larger letters indicate fully conserved residues. Statistically well-supported motif mimics (p < 10–3) were identified via the Eukaryotic Linear Motif resource (http://elm.eu.org). Those predicted to be involved in intracellular trafficking (*) and nuclear translocation (**) are shown in blue. Bars indicate location and length, and consensus sequences are boxed. NLS, nuclear localization signal. NRBOX, nuclear receptor box (or LxxLL) motif confers binding to nuclear receptors and is present in the C-terminal domain of all VM protein paralogs [including the natural L. interrogans deletion variant, Q72N53 (LA0591 cluster ID)]. Q72UG2 (LA3490) contains an additional LxxLL motif depicted as NRBOXa. E2F, LxxLFD motif characteristic of E2F family transcription factor. AP2a, DPF/W motif binds alpha and beta subunits of AP2 adaptor complex. F4E, variant YxxxxL motif mediates binding to the dorsal surface of eukaryotic translation initiation factor, eIF4E. dCOP, di-tryptophan motif predicted to mediate retrograde trafficking from Golgi to the endoplasmic reticulum.