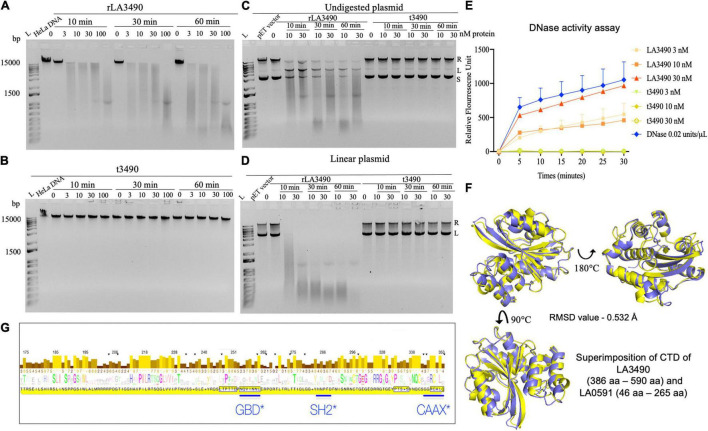
FIGURE 6.
DNase activity of leptospiral VM proteins. (A) DNase activity of rLA3490 observed upon incubation of 150 ng of DNA from HeLa cells in TM buffer containing 3 mM Mg2+ for indicated dose and time (absence of Mg+2 in reaction yielded no DNA degradation). Samples were subjected to 1% agarose gel electrophoresis. The DNase activity rLA3490 is indicated by smearing and disappearance of DNA; t3490 had no such effect. (B) Other recombinant VM proteins (LA0620, LA1400, LA1402, and LA0591) all had similar DNase activity (not shown). (C) DNase activity of rLA3490 on 400 ng of undigested plasmid pET28 shows partial degradation with uncoiling, linearizing, and partial degradation, and unaffected by t3490, shown by the white arrow. (D) DNase activity of rLA3490 on linearized plasmid shows complete disappearance of linear and relaxed plasmid, with dose- and time-dependent smearing. L, DNA ladder. (E) Quantification of rLA3490 DNase activity using real-time PCR and a FAM fluorescence probe. Bovine DNase, 0.02 U/μl, was used as positive control. Data represent the mean ± SD of three independent experiments. (F) Superimposition of AlphaFold generated CTD of LA3490 and LA0591, respectively. While LA3490 represents the vast majority of VM proteins with two RBLs, a CTD, and intervening functional sequences as visualized in Figure 1A, LA0591 lacks RBL1 and RBL2 but contains the rest of the functional sequences. This paralog represented by LA0591 is only fully present in L. interrogans species but not in other pathogenic Group I Leptospira. The CTDs of LA3490 and LA0591 are predicted to be highly conserved at the structural level despite amino acid sequence divergence, as shown by the RMSD values of 0.532 Å. (G) Eukaryotic trafficking motifs are present in the DNase-containing CTDs of leptospiral VM protein paralogs. GBD, GTPase-binding domain ligand, which is an amphipathic α-helix found in the C-terminal VCA segment of WASP/N-WASP proteins. SH2, Src Homology 2 domain, which is a broadly conserved protein interaction module central to tyrosine kinase signaling, and found in ubiquitin ligases, transcription factors, and guanine nucleotide exchange factors. The canonical CAAX motif is found in most Leptospira VM protein paralogs (but neither Q72UG2 nor Q72N53).