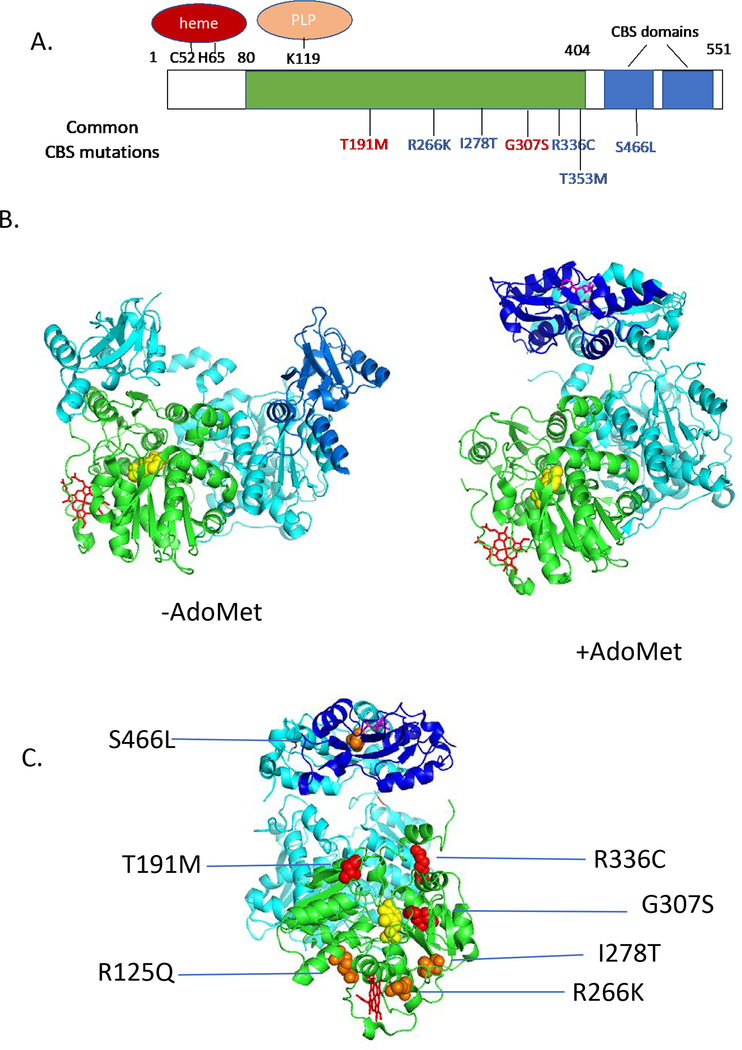
Figure 1.
Human CBS structure and mutations. A. One-dimensional drawing showing homology domains of CBS. Green shows region similar to other PLP-containing enzymes, blue shows CBS domains. Above the rectangle shows location of key residues binding heme and PLP. Below show some patient derived mutations. Red mutations are “non-rescuable”, while blue mutations are rescuable. B. Structure of human CBS (Δ516–525) as determined in the presence and absence of AdoMet. One subunit of the dimer is shaded as in A., while the other is shaded in teal. PLP is shown in yellow, heme in red, and AdoMet in magenta. C. Patient derived mutants mapped onto AdoMet activated structure (rotated 90° on vertical axis compared to image in A.). Red mutations are non-responsive, while orange are pyridoxine-rescuable.