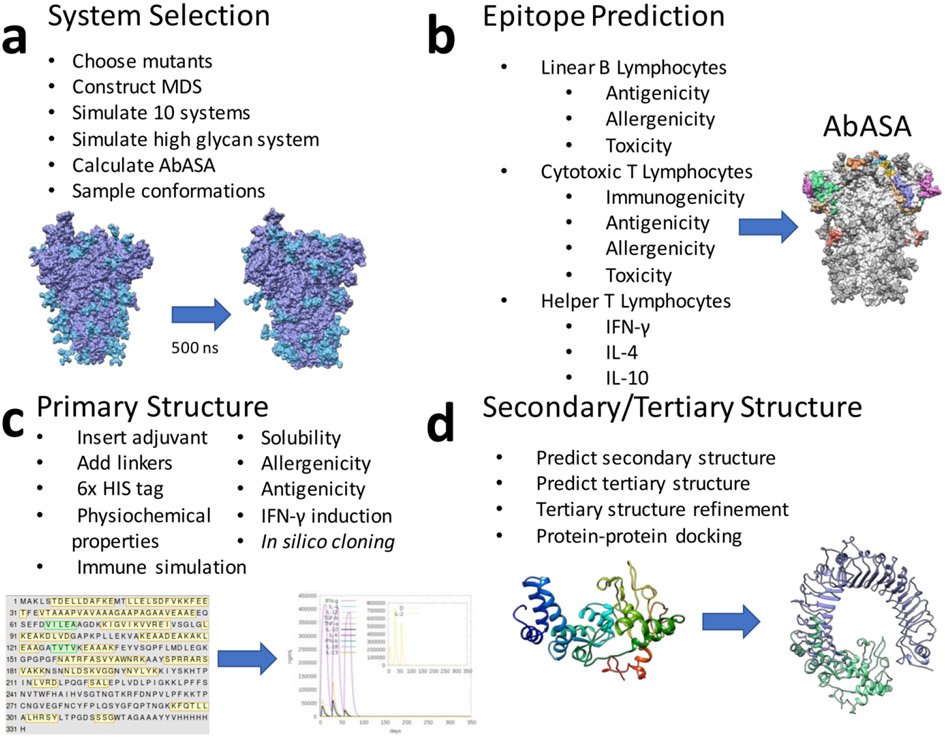
Figure 1: Overall workflow for project.
a) Selection of the systems used to generate conformations to be used in the linear B lymphocyte prediction. b) Epitope predictions, including linear B lymphocyte, cytotoxic T lymphocyte, and helper T lymphocytes. Predicted epitopes were assessed for multiple immune-relevant properties, as well as their ability to be accessed by a simulated antibody (antibody accessible surface area, AbASA). c) The final selected epitopes were linked together, along with an N-terminal adjuvant, using EAAAK, GPGPG, AAY, and KK linkers. This sequence was assessed for immune-relevant properties and simulated immune response. d) The secondary and tertiary structures were predicted and refined, and the final 3D structure was docked using protein-protein docking to toll-like receptor 2 and 4 (TLR2/4).