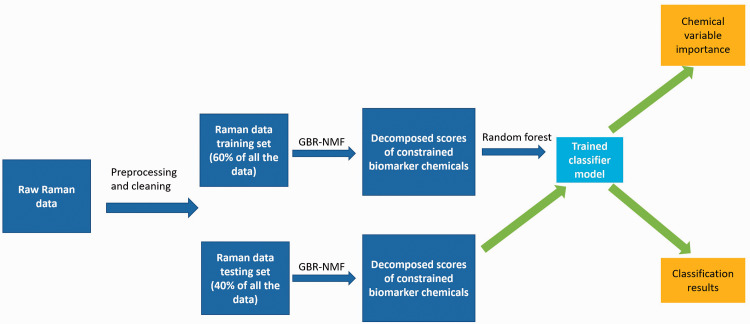
Figure 1.
GBR-NMF-RF data analytical framework workflow. After the cellular spectra were processed, the data set was split into 10 training and testing sets pairs. GBR-NMF dimension reduction was performed individually on the training set. The GBR-NMF decomposed chemical scores from the training set were used to random forests (RFs) based on the molecular histotype information of the cellular data. The GBR-NMF decomposed chemical scores from the testing set were used to get the classification performance and variable importance.