Table 5. Stages in the evolution of the SDPD of DCQ for the structure that finally turned out to be the correct one.
The initial random trial structure (GO1–GO2) in P1 evolved through the fitting steps of the global optimization run (GO3–GO4) and went through automatic cell transformation and re-evaluation fine fit (RE). It was then transformed to
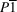 and subjected to the final user-controlled Rietveld refinement (UR). The similarity values S
12(l) refer to the comparison of the simulated pattern to the background-corrected experimental pattern based on the 2θ comparison range set for the global optimization.
and subjected to the final user-controlled Rietveld refinement (UR). The similarity values S
12(l) refer to the comparison of the simulated pattern to the background-corrected experimental pattern based on the 2θ comparison range set for the global optimization.
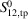 refers to the comparison of the powder pattern simulated by TOPAS and the experimental pattern. E
FF denotes the force-field energy.
refers to the comparison of the powder pattern simulated by TOPAS and the experimental pattern. E
FF denotes the force-field energy.
| Random | Fast raw fit | Better fit 1 | Better fit 2 | Automatic transformation | Re-evaluation fit | Final Rietveld refinement | ||
|---|---|---|---|---|---|---|---|---|
| GO2 | GO3 | GO4 | GO4 | RE1 | RE2 | UR | ||
| Space group (Z) | P1 (Z = 1) | P1 (Z = 1) | P1 (Z = 1) | P1 (Z = 1) | P1 (Z = 1) | P1 (Z = 1) |
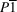 (Z = 1)
(Z = 1) |
|
| l (°) | 1.0 | 0.72 | 0.58 | 0.43 | 0.1 | |||
| Similarity S 12(l) | 0.7331 | 0.8997 | 0.9615 | 0.9536 | 0.9714 | |||
| Reference similarity |
|
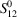
|
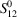
|
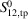
|
||||
| 0.9186 | 0.9836 | 0.9868 | 0.9969 | |||||
| V/Z (Å3 mol−1) | 355.07 | 388.16 | 388.93 | 389.58 | 389.58 | 391.08 | 377.20 | |
| a (Å) | 3.868 | 3.797 | 3.797 | 3.797 | 3.797 | 3.797 | 3.772 | |
| b (Å) | 5.888 | 6.442 | 6.508 | 6.521 | 6.521 | 6.551 | 6.479 | |
| c (Å) | 15.944 | 16.233 | 16.132 | 16.131 | 16.131 | 16.125 | 15.774 | |
| α (°) | 85.99 | 86.53 | 85.30 | 85.04 | 94.96 | 95.33 | 93.74 | |
| β (°) | 90.85 | 90.74 | 91.14 | 91.14 | 91.14 | 91.14 | 92.19 | |
| γ (°) | 78.68 | 78.41 | 78.41 | 78.41 | 101.59 | 101.49 | 100.92 | |
| Δφ x (°) | −15.9 | −15.8 | −24.3 | −25.2 | R exp (%) | 4.599 | ||
| Δφ y (°) | −66.5 | −66.3 | −64.7 | −64.5 | R wp (%) | 5.219 | ||
| Δφ z (°) | 24.6 | 25.0 | 34.4 | 35.2 | R wp′ (%) | 8.438 | ||
| E FF (kJ mol−1) | −115.3 | −207.7 | −207.4 | GoF | 1.135 | |||
