Abstract
Mass casualty exposure scenarios from an improvised nuclear device are expected to be far more complex than simple photons. Based on the proximity to the explosion and potential shielding, a mixed field of neutrons and photons comprised of up to approximately 30% neutrons of the total dose is anticipated. This presents significant challenges for biodosimetry and for short-term and long-term medical treatment of exposed populations. In this study we employed untargeted metabolomic methods to develop a biosignature in urine and serum from C57BL/6 mice to address radiation quality issues. The signature was developed in males and applied to samples from female mice to identify potential sex differences. Thirteen urinary (primarily amino acids, vitamin products, nucleotides) and 18 serum biomarkers (primarily mitochondrial and fatty acid β oxidation intermediates) were selected and evaluated in samples from day 1 and day 7 postirradiation. Sham-irradiated groups (controls) were compared to an equitoxic dose (3 Gy X-ray equivalent) from X rays (1.2 Gy/min), neutrons (~1 Gy/h), or neutrons-photons. Results showed a time-dependent increase in the efficiency of the signatures, with serum providing the highest levels of accuracy in distinguishing not only between exposed from non-exposed populations, but also between radiation quality (photon exposures vs. exposures with a neutron component) and in between neutron-photon exposures (5, 15 or 25% of neutrons in the total dose) for evaluating the neutron contribution. A group of metabolites known as acylcarnitines was only responsive in males, indicating the potential for different mechanisms of action in baseline levels and of neutron-photon responses between the two sexes. Our findings highlight the potential of metabolomics in developing biodosimetric methods to evaluate mixed exposures with high sensitivity and specificity.
INTRODUCTION
Nuclear terrorism and the potential for wartime use of nuclear weapons remain a high national security issue. Nuclear proliferation around the world has led to the necessity to develop effective and rapid methods to assess thousands of individuals in the event of a detonation. Significant efforts in the field of biodosimetry have produced numerous biomarkers and bioindicators of exposure, although efforts have mainly concentrated on single-exposure scenarios, e.g., acute doses of photons, and utilizing minimally invasive methods for sample acquisition (1–10). However, the environment of a detonation from an improvised nuclear device (IND) will create a far more complex scenario, containing mixed fields of neutrons and photons delivered acutely and subsequent low-dose-rate photon exposures due to groundshine and ingested/inhaled radionuclides, with the relative contribution of these fields depending on the proximity to the explosion, shielding materials and rapidity of evacuation (11–13). Measurements of the physical dose may lead to misinterpretation and administration of the wrong countermeasures, while the biological dose may actually be higher than initial readings indicate. It is therefore imperative to develop biodosimetric methods that could effectively distinguish between different mixed fields based on biological parameters.
Dosimetry, radiation weighing factors and relative biological effectiveness factors (RBEs) of the atomic bomb survivors are constantly being evaluated with new criteria (14, 15). A constant RBE of 10 was previously utilized by the Radiation Effects Research Foundation (RERF) for neutrons (16); however, taking into account that most exposures are mixed field, it was determined that the RBE depends on both neutrons and photons, with particular emphasis on the photon component (16). In addition, new models indicate that an RBE of 10 is a significant underestimation and therefore organ-specific-RBE values are much higher (14). Cordova and Cullings also indicated that future directions should include estimation of the RBE as a function of sex, based on recently reported data from the Life Span Study (LSS) (14, 17). Therefore, mixed-field exposures remain complex in their interpretation, particularly in the context of biological responses, and can lead to different and unexpected end points compared to pure photons.
Initial research through transcriptomics (18–21) and lipidomics (22), for the purposes of biodosimetry, has led to the identification of biomarkers that could be used for triage. Through transcriptomics, researchers identified neutron percentage-dependent responses in human peripheral blood cells (21), particularly in TP53-regulated genes. The same group also identified specific pathways in mouse experiments, such as protein translation, that were suppressed in groups that contained a neutron component (20). Lipidomic analysis surprisingly identified higher pro-inflammatory lipid-based responses in mixed neutron-photon fields, increasing with higher neutrons in the total dose, and with synergistic responses in certain lipid classes in the mixed field (22). The mixed neutron-photon fields and neutron spectra in these studies were designed to closely resemble the Hiroshima neutron spectrum and 1.5 km from the detonation (11, 23, 24) at the Columbia University accelerator neutron irradiation facility (Radiological Research Accelerator Facility; RARAF, Irvington, NY). This distinction is important from other neutron facilities which utilize fission spectra and may be designed specifically for protracted exposures (25–27).
We previously utilized metabolomics in urine and serum to qualitatively assess differences between single exposures of neutrons or photons (28), with emphasis on amino acid metabolism and fatty acid β oxidation perturbations. We expand our objectives to investigate metabolic responses in neutron-photon exposures with varying neutron percentages (5–25%) to a total dose of 3 Gy. The goal of this study was to develop unique small molecule signatures in urine and serum to not only differentiate between complex exposures, pure photons, and unexposed populations, but also to effectively differentiate between more-complex-field exposures with different neutron contributions to a constant total dose. The signatures were initially developed in male mice and tested in female mice to evaluate sex-independent responses and were evaluated at two time points pertinent to radiation biodosimetry. To our knowledge, this is the first study to evaluate neutron-photon metabolomic responses, as could be encountered from an IND explosion, in biofluids utilizing both male and female cohorts, and design biosignatures that can further address the complexity of such exposures. However, it should be noted that extrapolation to human populations for mixed-field irradiations is not feasible at the moment and will require further assessment with appropriate samples.
MATERIALS AND METHODS
Chemicals
All chemicals and solvents for untargeted metabolomics and tandem mass spectrometry (MS/MS) of the identified biomarkers were of the highest purity. 2-Hydroxybutyric acid, azelaic acid (nonanedioic acid), docosahexaenoic acid, taurine, acetylcarnitine, carnitine, creatine, eicosapentaenoic acid, glucose, guanosine, linoleylcarnitine, oleoylcarnitine, palmitoylcarnitine, phenylalanine, phytosphingosine, tetradecanoylcarnitine, valine, xanthine, 4-pyridoxic acid, cis-aconitic acid, erythronic acid, glutamic acid, lysine, uridine, xanthosine, pipecolic acid, nicotinic acid, pantothenic acid (vitamin B5), pyridoxal (vitamin B6), thymidine, thymine, 4-nitrobenzoic acid, chlorpropamide, and debrisoquine sulfate were obtained from Sigma-Aldricht LLC, (St. Louis, MO).
Animal Irradiations, Dosimetry and Sample Collection
Irradiations and dosimetry were conducted at Columbia University as described elsewhere (22) in detail, including doses for neutrons and photons. Briefly, C57BL/6 male and female mice (n = 5 per group) were purchased from Charles River Laboratories (Wilmington, MA) at 8–10 weeks old. Neutron irradiations (5, 15 or 25% of the total dose; dose rate ~1 Gy/h) were followed immediately with X-ray irradiations (250 kVp; HVL 2 mm Cu, 1.2 Gy/min) to a total dose of 3 Gy. Neutron dosimetry was performed on the day of irradiation as reported elsewhere (24). A custom tissue-equivalent (TE) gas ionization chamber (29) was first calibrated using a NIST-traceable 226Ra source and then irradiated in the same geometry as the mice, giving total dose rate (neutrons plus concomitant photons). A compensated Geiger-Mueller dosimeter, which has very low sensitivity to neutrons, was irradiated in the same geometry and used to measure the photon dose rate. These two dose rates were used to calibrate an independent monitor chamber, placed at a different angle from the mice and used to control irradiations. X-ray dosimetry was performed using a commercial NIST-traceable ionization chamber (Radcal® 10X6–6; Monrovia, CA) calibrated to air kerma. Six groups were included in the experiment, including sham (controls), neutrons alone (0.75 Gy neutrons with 0.16 Gy concomitant c rays; equitoxic to 3 Gy of X rays), X rays (3 Gy), 5% neutrons in total dose, 15% neutrons in total dose, and 25% neutrons in total dose (22). Spot urine samples and serum from cardiac punctures were collected at the time of euthanasia (day 1 or 7 postirradiation), flash frozen, and stored at –80°C until shipment to Georgetown University (Washington, DC).
Sample Preparation, Data Analysis and Metabolite Identification
Metabolite extraction from urine and serum is described elsewhere (28). Untargeted metabolomic analysis was conducted through ultra-performance liquid chromatography (UPLC) coupled to time-of-flight mass spectrometry (Xevo G2 or Xevo G2S; Waters® Corp., Milford, MA) (Supplementary Table S1; https://doi.org/10.1667/RADE-20-00032.1.S1). Chromatographic deconvolution and peak analysis was performed using the software Progenesis QI (NonLinear Dynamics, Newcastle, UK). Quality controls (QCs) from pooled samples were run every 10 samples for assessment of chromatographic quality and one of the QCs automatically picked by the software was used for peak alignment. All data were acquired with the MSE function that collects fragmentation patterns for the majority of ions. Urinary normalization was performed to creatinine levels [M + H]+ = 114.0667, while serum normalization was performed with the function ‘‘Normalize to all compounds’’ through Progenesis QI. Creatinine P values were determined with Kruskal-Wallis test and Dunn’s multiple comparison testing for two group comparisons. The only exception occurred for male urine day 1 data that was normalized to all compounds. Putative identities were assigned through the databases METLIN (30) (matched to empirical fragmentation data), LIPID MAPS (31) and HMDB (32).
Potential biomarkers were determined through the software MetaboAnalyst 4.0 (33). Briefly, features with > 75% missing values were removed and data were Pareto scaled. Statistically significant ions per time point were determined through one-way analysis of variance (ANOVA) with Tukey’s post hoc analysis and an adjusted P value (false discovery rate; FDR) cutoff of 0.05. Select ions with P < 0.05 were further positively identified through MS/MS, with fragmentation patterns and retention time in a QC for each candidate matched to those from a pure standard.
Biosignature Development
Biomarkers with P < 0.05, as determined through ANOVA testing, were initially identified in male mouse data and were further examined in female mice for determination of sex differences or similarities in the responses. Biomarkers in female samples that were below sensitivity detection in Progenesis QI were extracted using the software TargetLynx (Waters Corp.). Subsequent analyses were conducted based on the identified biomarkers with the software MetaboAnalyst 4.0 (33). Outlier testing for each metabolite was conducted through GraphPad Prism version 6 (La Jolla, CA) with the ROUT method (Q = 1%) and were removed from subsequent analysis. Pathway enrichment based on the validated metabolites identified in male samples was shown in a bar chart format. Sparse partial least square discriminant analysis (sPLS-DA) (34) of the first two components with limited variables and a fivefold CV validation method were constructed without removal of missing values and replacement with one half of the minimum value from the original data and Pareto scaled. Multivariate receiver operating characteristic (ROC) curves and area under the curve (AUC) values were constructed for each signature and calculated using MetaboAnalyst based on the Random Forests algorithm for performance evaluation of the signature and classification and feature ranking method. All neutron-containing samples (pure neutrons or neutron-photon groups) were compared to X rays. ROC curves were combined to demonstrate the effect of complex exposures in distinguishing between two types of radiation exposure scenarios with high sensitivity and specificity. AUCs closer to 1 indicated an excellent degree of separability between the groups based on the biosignature.
Graphical Representation and Statistical Analysis
Graphical representation was conducted using GraphPad Prism version 6 software, which also allowed for statistical analysis. Analysis within multiple groups was conducted with one-way ANOVA and Tukey’s multiple comparison testing (P < 0.05 considered significant), while analysis within two groups was conducted with a parametric two-tailed t test with Welch’s correction (P < 0.05 considered significant). Fold changes were calculated as log2 compared to the control group.
RESULTS
Candidate biomarkers in urine and serum samples were identified based on an ANOVA FDR-corrected P value of <0.05 among all groups in either day 1 or day 7 (Table 1) and further validated through MS/MS. Normalization in urine was performed to all compounds for male day 1 (creatinine P < 0.0001) and creatinine for male day 7 (P = 0.0157; however, Dunn’s testing showed no statistically significant P values between two group comparisons) and female day 1 and day 7 (P > 0.05 for both). Since different normalization methods were used, fold changes are used as the best method to assess the directionality of the changes (Tables 2 and 3). A total of 13 urinary and 18 serum metabolites were selected to develop a signature in male mice and tested in female mice for identification of sex differences or similarities (Table 1). The urinary signature was comprised of: 4-pyridoxic acid, cis-aconitic acid, erythronic acid, glutamic acid, lysine, uridine, xanthosine, pipecolic acid, nicotinic acid (vitamin B3), pantothenic acid (vitamin B5), pyridoxal (vitamin B6), thymidine and thymine. The serum signature was comprised of: 2-hydroxybutyric acid, azelaic acid (nonanedioic acid), docosahexaenoic acid, taurine, acetylcarnitine, carnitine, creatine, eicosapentaenoic acid, glucose, guanosine, linoleylcarnitine, oleoylcarnitine, palmitoylcarnitine, phenylalanine, phytosphingosine, tetradecanoylcarnitine, valine and xanthine. Pathway enrichment, to assign biological significance to the biomarkers, showed distinctly different pathway involvement of the biomarkers in each of the two biofluids (Fig. 1). In particular, urinary biomarkers included primarily amino acids, nucleotides, and vitamin products, whereas serum biomarkers were dominated by mitochondrial and fatty acid b oxidation intermediates, among others.
TABLE 1.
MS/MS Verified Metabolites in Murine Serum and Urine
| Validated metabolite | FDR-corrected
P value |
|||||||
|---|---|---|---|---|---|---|---|---|
| Day 1 | Day 7 | m/z observed | m/z expected | ppm error | Retention time | Adduct | ||
| Serum | ||||||||
| 1 | 2-Hydroxybutyric acid | NS | 0.04 | 103.0396 | 103.0401 | 4.85 | 0.48 | [M − H] − |
| 2 | Azelaic acid (nonanedioic acid) | NS | 0.03 | 187.0965 | 187.0976 | 5.88 | 4.22 | [M − H] − |
| 3 | Docosahexaenoic acid | NS | 0.03 | 327.2326 | 327.233 | 1.22 | 8.2 | [M − H] − |
| 4 | Taurine | NS | 0.004 | 124.0065 | 124.0074 | 7.26 | 0.32 | [M − H] − |
| 5 | Acetylcarnitine | 0.04 | NS | 204.1237 | 204.123 | 3.43 | 0.33 | [M + H]+ |
| 6 | L-carnitine | 0.05 | NS | 162.1131 | 162.1125 | 3.70 | 0.33 | |
| 7 | Creatine | 0.001 | 0.004 | 132.0772 | 132.0768 | 3.03 | 0.32 | [M + H]+ |
| 8 | Eicosapentaenoic | NS | 0.04 | 303.2326 | 303.2319 | 2.31 | 8.04 | [M + H]+ |
| 9 | acid Glucose | 0.05 | 0.03 | 203.0533 | 203.0526 | 3.45 | 0.3 | [M + H]+ |
| 10 | Guanosine | 0.05 | NS | 306.0814 | 306.0809 | 1.63 | 0.38 | [M+Na]+ |
| 11 | Linoleylcarnitine | NS | 0.004 | 424.3424 | 424.3421 | 0.71 | 7.13 | [M + H]+ |
| 12 | Oleoylcarnitine | NS | 0.03 | 426.3579 | 426.3579 | 0.00 | 7.35 | [M + H]+ |
| 13 | Palmitoylcarnitine | NS | 0.004 | 400.3422 | 400.3421 | 0.25 | 7.27 | [M + H]+ |
| 14 | L-Phenylalanine | NS | 0.00 | 166.08473 | 166.0863 | 9.45 | 0.3 | [M + H]+ |
| 15 | Phytosphingosine | 1.46E–06 | 0.02 | 318.3006 | 318.3003 | 0.94 | 6.74 | [M + H]+ |
| 16 | Tetradecanoylcarnitine | NS | 0.01 | 372.3114 | 372.3108 | 1.61 | 6.91 | [M + H]+ |
| 17 | L-valine | NS | 0.04 | 140.0688 | 140.0682 | 4.28 | 0.3 | [M+Na]+ |
| 18 | Xanthine | 0.02 | 0.002 | 153.0415 | 153.0407 | 5.23 | 0.38 | [M + H]+ |
| Urine | ||||||||
| 1 | 4-Pyridoxic acid | NS | 0.0003 | 182.045 | 182.0459 | 0.67 | 0.45 | [M − H] − |
| 2 | cis-Aconitic acid | NS | 0.01 | 173.0082 | 173.0092 | 0.62 | 0.38 | [M − H] − |
| 3 | Erythronic acid | NS | 0.002 | 135.029 | 135.0299 | 0.57 | 0.33 | [M − H] − |
| 4 | L-glutamic acid | 0.02 | NS | 146.045 | 146.0459 | 0.59 | 0.35 | [M − H] − |
| 5 | L-lysine | 0.04 | NS | 145.0973 | 145.0983 | 0.55 | 0.3 | [M − H] − |
| 6 | Uridine | NS | 0.0005 | 243.0612 | 243.0623 | 0.57 | 0.33 | [M − H] − |
| 7 | Xanthosine | NS | 0.03 | 283.0675 | 283.0684 | 0.69 | 0.47 | [M − H] − |
| 8 | L-pipecolic acid | 0.01 | 0.01 | 130.0869 | 130.0863 | 4.61 | 0.35 | [M + H]+ |
| 9 | Nicotinic acid | 7.64E–06 | NS | 124.04 | 124.0393 | 5.64 | 0.74 | [M + H]+ |
| 10 | Pantothenic acid | 0.002 | 0.01 | 220.1186 | 220.1179 | 3.18 | 0.74 | [M + H]+ |
| 11 | Pyridoxal (vitamin B6) | 0.02 | 0.01 | 168.0643 | 168.0655 | 7.14 | 0.66 | [M + H]+ |
| 12 | Thymidine | NS | 0.01 | 265.0814 | 265.0795 | 7.17 | 0.55 | [M+Na]+ |
| 13 | Thymine | NS | 0.003 | 127.0508 | 127.0502 | 4.72 | 0.35 | [M + H]+ |
NS = not significant.
TABLE 2.
Log2 Fold Changes in Urine
| X rays |
Neutrons |
5% |
15% |
25% |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Male | Female | Male | Female | Male | Female | Male | Female | Male | Female | |
| Day 1 | ||||||||||
| 4-Pyridoxic acid | −0.27 | −1.70 | −0.28 | −1.46 | 0.07 | −1.47 | −0.22 | −1.52 | 0.16 | −1.24 |
| cis-Aconitic acid | 0.43 | 0.13 | 0.59 | 0.20 | 0.22 | 0.39 | 0.36 | 0.02 | 0.21 | 0.27 |
| Erythronic acid | 0.66 | −1.82 | 0.64 | −1.78 | 0.26 | −1.73 | 0.52 | −2.08 | 0.29 | −1.87 |
| L-glutamic acid | 0.83 | −3.42 | 0.72 | −3.15 | 0.34 | −3.10 | 0.42 | −3.57 | 0.36 | −3.23 |
| L-pipecolic acid | −0.53 | −0.56 | −0.60 | −0.49 | −0.62 | −0.37 | −0.43 | −0.40 | −0.65 | −0.45 |
| Lysine | 0.56 | −0.99 | 0.91 | −0.65 | −0.12 | −0.99 | 0.29 | −0.66 | 0.20 | −0.83 |
| Nicotinic acid | −0.50 | −7.33 | −0.47 | −7.52 | −0.42 | −7.26 | −0.35 | −7.49 | −0.47 | −6.94 |
| Pantothenic acid | −0.39 | −0.55 | −0.31 | −0.50 | −1.06 | 0.11 | −0.98 | −0.29 | −0.98 | 0.06 |
| Pyridoxal | −0.59 | −0.25 | −0.52 | 0.02 | −0.52 | −0.81 | −0.33 | −0.50 | −0.32 | −0.27 |
| Thymine | −0.47 | −1.31 | −0.39 | −1.63 | −0.43 | −1.21 | −0.36 | −1.38 | −0.52 | −1.39 |
| Uridine | 0.45 | −2.10 | 0.33 | −2.08 | 0.18 | −1.99 | 0.24 | −2.33 | 0.18 | −2.01 |
| Xanthosine | 0.56 | −4.51 | 0.46 | −4.42 | 0.80 | −4.25 | 0.60 | −4.77 | 0.87 | −4.15 |
| Thymidine | 0.3 | −2.49 | 0.19 | −2.58 | 0.42 | −2.20 | 0.57 | −2.50 | 0.40 | −2.26 |
| Day 7 | ||||||||||
| 4-Pyridoxic acid | 0.13 | −0.58 | 0.47 | −0.61 | 0.58 | 0.28 | 1.05 | −0.12 | −0.23 | 0.04 |
| cis-Aconitic acid | −0.03 | 0.02 | 0.08 | −0.26 | 0.26 | −0.04 | 0.44 | 0.04 | 0.16 | 0.13 |
| Erythronic acid | 0.17 | 0.15 | 0.03 | −0.46 | 0.13 | −0.21 | 0.30 | −0.26 | 1.19 | −0.12 |
| L-glutamic acid | −0.02 | −0.21 | 0.33 | −0.61 | 0.36 | −0.22 | 0.64 | −0.44 | 0.35 | −0.30 |
| L-pipecolic acid | −0.18 | −0.43 | −0.21 | −0.34 | 0.07 | 0.27 | −0.03 | −0.10 | −0.05 | 0.00 |
| Lysine | 0.30 | 0.28 | 0.18 | 1.14 | 0.20 | 0.99 | 0.53 | 1.61 | −0.98 | 0.65 |
| Nicotinic acid | −0.37 | −0.61 | −0.11 | −0.09 | −0.36 | −0.14 | −0.04 | −0.07 | −0.06 | −0.07 |
| Pantothenic acid | 0.28 | −0.57 | 0.47 | −0.32 | 0.23 | 0.14 | 0.29 | −0.09 | 0.29 | 0.17 |
| Pyridoxal | −0.07 | −0.09 | −0.22 | 0.25 | 0.07 | −0.09 | 0.07 | 0.05 | 0.21 | −0.29 |
| Thymine | −0.57 | −0.18 | −0.38 | −0.44 | −0.08 | −0.27 | −0.16 | −0.48 | −0.27 | −0.14 |
| Uridine | 0.08 | −0.16 | 0.10 | −0.52 | 0.09 | −0.12 | 0.36 | −0.34 | 0.70 | −0.27 |
| Xanthosine | −0.09 | −0.70 | −0.18 | −0.70 | −0.24 | 0.02 | 0.32 | −0.58 | −0.51 | −0.40 |
| Thymidine | 0.08 | −0.34 | 0.33 | −0.35 | 0.40 | 0.07 | 0.39 | −0.16 | 0.15 | −0.03 |
TABLE 3.
Log2 Fold Changes in Serum
| X rays |
Neutrons |
5% |
15% |
25% |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Male | Female | Male | Female | Male | Female | Male | Female | Male | Female | |
| Day 1 | ||||||||||
| Acetylcarnitine | −0.69 | 0.15 | −0.80 | 0.12 | −1.20 | 0.45 | −0.86 | 0.40 | −0.81 | −0.13 |
| Carnitine | −0.56 | 0.01 | −0.68 | 0.04 | −1.07 | 0.17 | −0.76 | 0.41 | −0.71 | −0.15 |
| Creatine | −0.50 | 0.06 | −0.72 | 0.37 | −0.75 | 0.17 | −0.91 | 0.02 | −1.05 | 0.60 |
| Eicosapentaenoic acid | 0.68 | 2.09 | 2.15 | 1.60 | 0.30 | 2.38 | 0.18 | 1.98 | 0.42 | 1.50 |
| Glucose | −0.48 | −0.33 | −0.62 | 0.15 | −0.96 | 0.15 | −0.75 | 0.02 | −0.69 | −0.27 |
| Guanosine | −2.70 | −0.99 | −6.71 | −0.27 | −5.74 | 0.12 | −4.81 | 0.03 | −5.66 | −0.04 |
| Linoleylcarnitine | −0.24 | 0.72 | 2.14 | 1.19 | 0.13 | 0.23 | −3.02 | 0.83 | −4.60 | 0.62 |
| Oleoylcarnitine | −0.31 | 0.47 | 1.72 | 1.05 | 0.16 | 0.38 | −3.04 | 0.86 | −4.24 | 0.51 |
| Palmitoylcarnitine | −0.47 | 0.55 | 1.41 | 0.65 | 0.22 | 0.45 | −1.64 | 0.12 | −2.51 | 0.56 |
| L-phenylalanine | −0.02 | −0.94 | 0.08 | −0.71 | −0.72 | −1.00 | −0.40 | −1.23 | −0.22 | −0.45 |
| Phytosphingosine | −2.26 | −0.004 | −2.54 | 0.005 | −1.16 | 0.03 | −1.08 | 0.05 | −1.19 | 0.05 |
| Tetradecanoylcarnitine | −0.77 | 0.40 | 1.15 | 0.63 | −0.67 | 1.04 | −4.42 | 0.66 | −4.27 | 0.20 |
| L-valine | −0.18 | −0.07 | −0.35 | 0.21 | −0.64 | 0.42 | −0.60 | 0.40 | −0.45 | 0.02 |
| Xanthine | −2.85 | - | −3.19 | 1.00 | −2.83 | 1.22 | −2.79 | −2.54 | −1.79 | −1.09 |
| 2-Hydroxybutyric acid | −0.17 | −0.94 | −0.42 | −0.47 | 0.21 | −0.38 | −0.05 | −0.25 | 0.20 | −0.82 |
| Azelaic acid | 0.25 | 0.23 | 0.29 | 0.37 | 0.41 | 0.35 | 0.53 | 0.13 | 0.31 | 0.24 |
| Docosahexaenoic acid | 0.47 | −0.83 | 0.93 | −0.50 | 0.23 | −0.58 | 0.28 | −0.60 | 0.08 | −0.95 |
| Taurine | −0.08 | 0.13 | −0.45 | 0.34 | −0.09 | 0.43 | −0.78 | 0.02 | −0.50 | 0.29 |
| Day 7 | ||||||||||
| Acetylcarnitine | −0.75 | 0.22 | −0.75 | −0.06 | −0.12 | 0.05 | −0.61 | 0.02 | −0.37 | −0.04 |
| Carnitine | −0.49 | −0.20 | −0.56 | −0.15 | 0.02 | −0.01 | −0.34 | −0.13 | −0.23 | −0.27 |
| Creatine | −0.45 | −0.80 | −0.85 | −0.50 | −0.70 | −0.02 | −1.05 | −0.29 | −0.99 | −1.11 |
| Eicosapentaenoic acid | 1.51 | 0.53 | 1.56 | 1.61 | 0.85 | 0.10 | 1.31 | 0.28 | 1.13 | 1.88 |
| Glucose | −0.65 | 0.14 | −0.91 | −0.01 | −0.06 | 0.48 | −0.40 | 0.18 | −0.27 | 0.44 |
| Guanosine | - | −0.63 | −4.74 | −0.50 | −5.91 | −0.27 | - | −0.87 | - | −0.80 |
| Linoleylcarnitine | −0.33 | 0.44 | −2.05 | −0.14 | −3.82 | −0.40 | −5.35 | −0.19 | - | 0.45 |
| Oleoylcarnitine | −0.38 | 0.50 | −3.03 | 0.41 | −4.72 | −0.85 | - | −0.40 | - | 0.04 |
| Palmitoylcarnitine | −0.26 | 0.03 | −1.60 | 0.06 | −2.62 | −0.52 | −4.11 | −0.44 | −6.73 | −0.32 |
| L-phenylalanine | −0.58 | −0.45 | −0.61 | 0.19 | 0.96 | 0.93 | 0.71 | 0.70 | 0.16 | −0.04 |
| Phytosphingosine | −0.77 | −0.05 | −0.70 | −0.09 | −0.42 | −0.08 | −0.01 | −0.14 | 0.01 | −0.06 |
| Tetradecanoylcarnitine | −0.49 | 0.29 | −2.14 | 0.08 | −2.49 | −0.27 | - | −0.33 | - | 0.08 |
| L-valine | −0.59 | −0.33 | −0.85 | −0.46 | 0.16 | −0.22 | −0.33 | −0.05 | −0.41 | −0.17 |
| Xanthine | −2.48 | −4.46 | −3.75 | −0.61 | −4.72 | - | −13.60 | - | −3.57 | −3.26 |
| 2-Hydroxybutyric acid | −4.23 | 0.01 | −0.95 | 0.66 | −0.26 | −0.33 | −0.49 | −0.11 | −1.87 | 0.31 |
| Azelaic acid | 1.30 | 0.20 | 0.21 | 0.46 | −0.14 | 0.20 | 0.05 | 0.01 | 0.79 | 0.52 |
| Docosahexaenoic acid | 0.94 | −0.08 | 0.26 | 0.27 | −0.01 | −0.19 | 0.18 | 0.16 | 0.96 | 0.04 |
| Taurine | −0.57 | −0.35 | −0.84 | −0.38 | −1.07 | −0.47 | −0.80 | −0.74 | −0.61 | −0.49 |
FIG. 1.
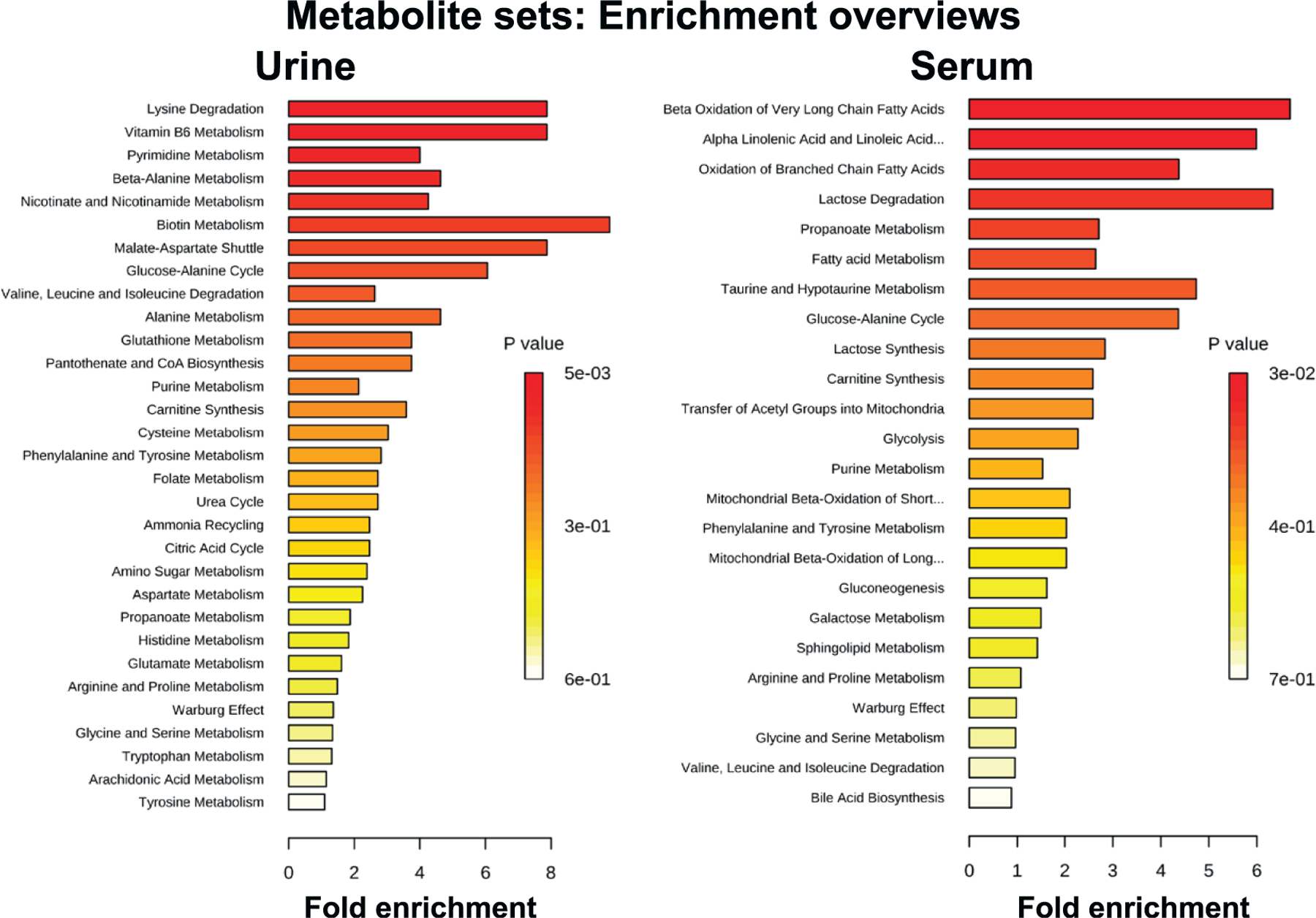
Pathway enrichment based on validated metabolites in urine and serum from male animals. Urine shows enrichment in polar metabolites, such as amino acids, while serum shows enrichment in metabolites associated with energy metabolism and mitochondrial involvement.
Log2 fold changes were calculated for each biomarker by comparing each irradiated group to controls in each time point (Tables 2 and 3). Changes at day 1 were variable between males and females in both urine and serum. Urinary alterations in biomarker levels from controls were immediate and more pronounced in females based on fold changes; however, more pronounced statistical significant changes were evident in males (Table 2; Supplementary Tables S2 and S4; https://doi.org/10.1667/RADE-20-00032.1.S2 and https://doi.org/10.1667/RADE-20-00032.1.S4, respectively). Erythronic acid, glutamic acid, uridine, xanthosine and thymidine were the only biomarkers that showed opposite responses between the two sexes (increased in males after irradiation, decreased in females). Responses at day 7 were more conservative and in the majority of the cases showed small changes from control levels (Table 2 and Supplementary Table S2). In the serum, however, augmented responses were observed at both time points that were investigated in either direction. The directionality of the changes in both sexes was more consistent at the later time point compared to the initial one. However, the males exhibited a far more dramatic response in individual metabolites compared to controls, unlike the females, where the responses were more dampened (Table 3; Supplementary Tables S3 and S5; https://doi.org/10.1667/RADE-20-00032.1.S3 and https://doi.org/10.1667/RADE-20-00032.1.S5, respectively).
Importantly, a select group of mitochondrial-related acylcarnitines (tetradecanoylcarnitine, oleoylcarnitine, palmitoylcarnitine and linoleylcarnitine) were the only biomarkers that could be utilized as stand-alone biomarkers to distinguish between radiation scenarios and control samples in male samples (Fig. 2). X rays alone did not show a statistically significant change from controls. However, all the neutron-irradiated groups showed an increased depletion in the serum with increasing neutron component in the total dose, reaching zero levels in some cases. Furthermore, responses in these biomarkers appear to be additive rather than synergistic. This is evident in the 25% group, which received an equal neutron dose of 0.75 Gy, as the ‘‘pure’’ neutron group. These dramatic responses in acylcarnitines, however, were not present in females, where levels of the metabolites were barely detectable with limited counts in each sample (less than 150) (Fig. 2), indicating a different mechanism of response. Acetylcarnitine and carnitine levels did not differ significantly between males and females in the overall responses to radiation, indicating that there are no significant differences between the two sexes in the carnitine shuttle mechanism, unlike the rest of the acylcarnitines.
FIG. 2.
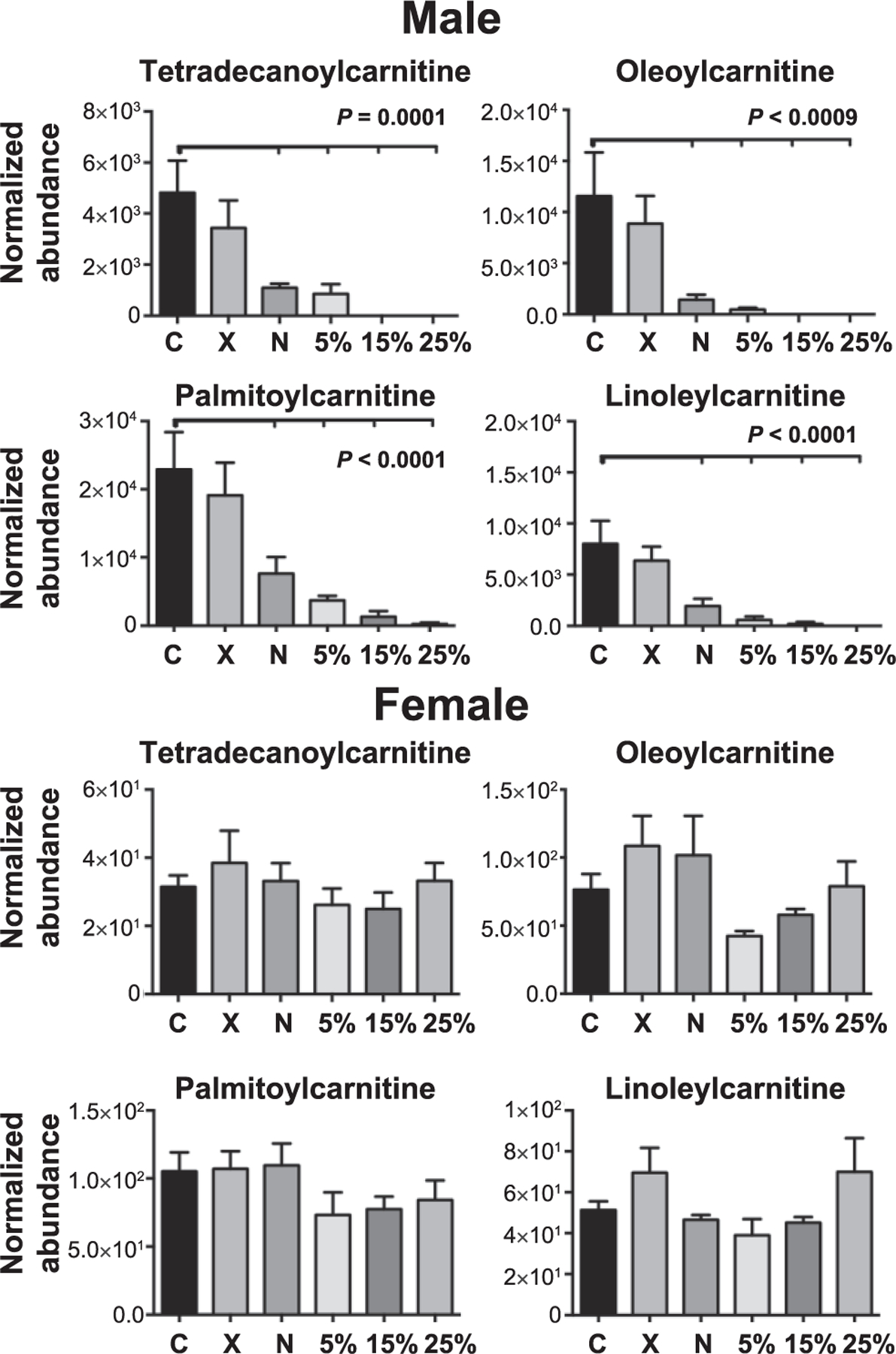
Levels of four acylcarnitines at 7 days postirradiation. Results in male serum samples show a gradual depletion, which are statistically significantly different from controls in the groups containing a neutron component. The levels of these acylcarnitines, on the other hand, are orders lower in female samples, with no significant changes from controls, demonstrating sex-dependent changes in neutron exposures and differential mechanisms of responses. P values were calculated using one-way ANOVA (statistical significance at P < 0.05), while tick marks reflect statistical significance compared to controls. All data presented are mean ± SEM
Combining the biomarkers in a signature further allowed for distinguishing between the different neutron-photon exposures. Utilizing sPLS-DA analysis (34) with 18 variables for serum and 13 for urine on each component, it was possible to distinguish between the three different neutron-photon irradiated groups with high predictive performance in both males and females (Fig. 3). In particular, component 1 was able to explain >40% of the variability in males and females on day 1, with less promising results in serum. However, 3D models incorporating components 1–3 showed distinct clustering between the groups (data not shown). Still, as time increased from the exposure, the clustering ability in both urine and serum, in males and females, also increased. As shown in Fig. 3, component 1 can explain >30% of the variability in all models, except in serum from females, which remains at 28.9%, with clustering further augmented in a 3D model (data not shown). In addition, combining all groups containing a neutron component as a single group and subsequently comparing control to X rays to neutrons, demonstrated the separation between groups based on the exposure and radiation quality, with responses increasing over time for both males and females, and more prominent in the serum (Supplementary Fig. S1; https://doi.org/10.1667/RADE-20-00032.1.S6).
FIG. 3.
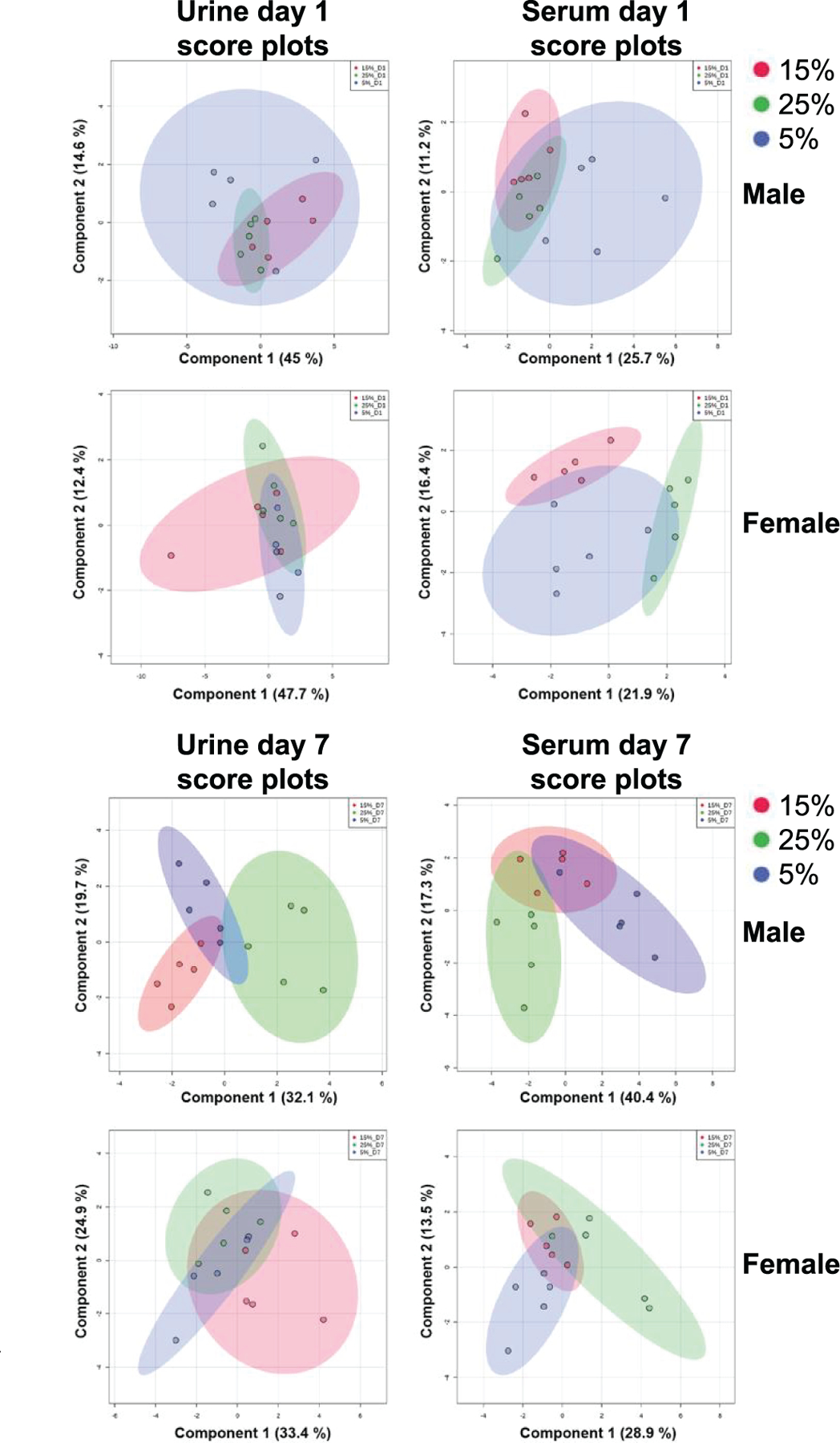
sPLS-DA score plots based on the urinary and serum signature developed in males demonstrate the effectiveness in separating the three neutron-photon groups with time dependence. Urine data utilized a combination of 13 metabolites, while serum data utilized a combination of 18 metabolites. Separation is shown for components 1 and 2 in all plots, with the variability in each component explained in percentage.
Finally, the signature was tested against the X-ray-irradiated group to evaluate the degrees of sensitivity and specificity in distinguishing between different radiation exposure scenarios. ROC curves were constructed for each neutron group (neutrons, 5%, 15% or 25%) and AUCs calculated using the software MetaboAnalyst version 4.0 (33) (Fig. 4). An equitoxic neutron dose was indistinguishable from X rays in the majority of the cases, except in urine from females at day 7 (AUC = 0.815), serum from females at day 1 (AUC = 0.875), and serum from males at day 7 (AUC = 0.965). High AUCs (>0.8) were calculated in urine for all except in females at day 1 (<0.8), indicating a high degree of confidence in distinguishing complex exposures from pure photons. Similar results were obtained in serum, except for females at day 7.
FIG. 4.
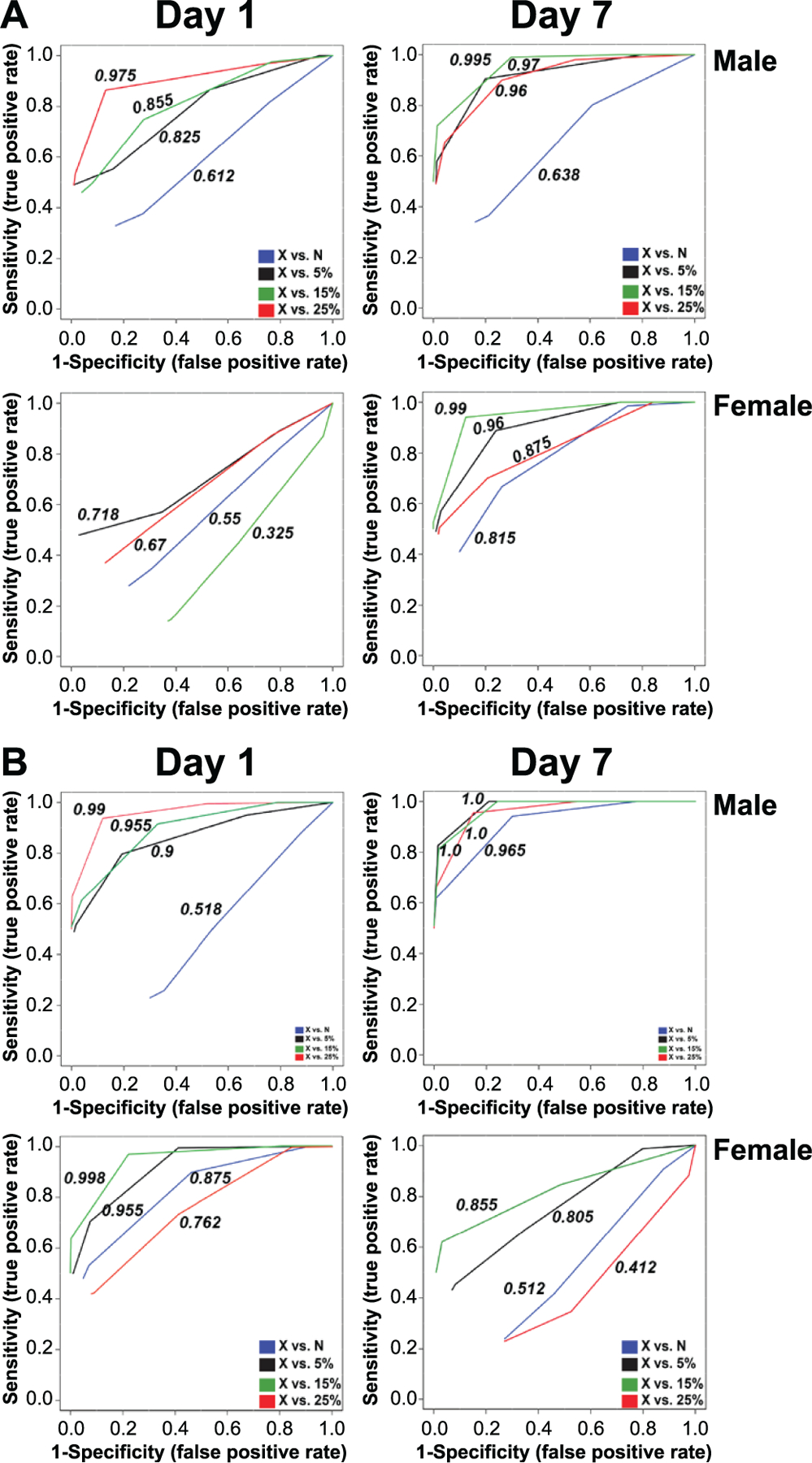
Superimposed ROC curves using Random Forests as a classification and feature ranking method, comparing neutrons and neutron-photon exposures to X rays. Each ROC curve shows the model averaged from all cross validation runs and the numbers represent the AUCs. Panel A. Urine analysis based on a biosignature of 13 metabolites from male samples extrapolated to females. Panel B. Serum analysis based on a biosignature of 18 metabolites from male samples extrapolated to females. X-axis: Specificity (false positive rate), Y-axis: Sensitivity (true positive rate). AUC >0.8 is considered a good model; AUC >0.9 is considered an excellent model.
DISCUSSION
The current study focused on developing metabolomics-based signatures in urine and serum from male mice that were exposed to a variety of neutron-photon scenarios for a total dose of 3 Gy. In addition, the validity of this sample was also tested in samples from female mice to determine whether a sex-independent signature could be developed. Our previously published study with lipidomics in male mice demonstrated complex exposure-dependent responses and the surprising synergistic effects between neutrons and photons, particularly with regards to inflammatory indicators (22). Transcriptomic studies utilizing an identical experimental setup and doses also identified significant responses based on neutrons compared to photons, however, fold changes of genes were not dependent of the percentage of neutrons (20). Such neutron-photon exposures will be encountered in the case of an IND explosion, where neutron contributions could potentially reach 25–30% of the total physical dose (35); however, the biological effects for acute adverse and stochastic end points will be much higher due to the high RBE associated with neutrons at a specific dose (14, 16). Questions therefore that need to be addressed in such a scenario include whether there was a neutron component in the total dose and what percentage of neutrons were present. Biodosimetry methods utilizing easily accessible biofluids (e.g., urine and serum, among others) have been investigated in an attempt to provide answers with high sensitivity and specificity. Metabolomics has been classified as a late-development phase study for human accidental exposure cases based on validated citrulline levels (9, 36). However, promising results can incorporate new biomarkers for the purposes of biodosimetry, further demonstrating the potential of metabolomics in clinical applications.
In the current study, a urinary signature including 13 metabolites and a serum signature including 18 metabolites were constructed based on responses observed in male mice. Polar metabolites (amino acids, nucleotides and vitamin products) were the primary types of metabolites identified in urine, while mitochondrial intermediates, such as acylcarnitines and fatty acid b oxidation-associated intermediates were part of the serum biomarkers. Fatty acid β oxidation products and associated genes have previously been identified as biomarkers distinguishing between neutron and photon exposures (18, 28). Both signatures were able to distinguish between controls and irradiated groups and provide separation between groups with a pure photon (3 Gy) and those containing a neutron component (neutrons equitoxic 0.75 Gy and neutrons-photons combined for a total of 3 Gy), therefore providing radiation quality-dependent biomarkers. However, as shown in Fig. 4, the specificity and sensitivity of the signature was dependent on the complex exposure and not on the neutron component alone, since X-ray vs. neutron AUCs were primarily low. Since the signature was developed in male mice, it performed with higher specificity and sensitivity in that cohort compared to females, as evident from the lower AUC values. Nonetheless, the predictive models showed high sensitivity and specificity in select female groups, e.g., day 7 X-ray irradiated vs. 5% or X-ray irradiated vs. 25%, and further enrichment of the signature with future female-specific biomarkers will make it possible to improve the biodosimetric signatures. Development of such signatures therefore is a dynamic process. Importantly, while responses at the early time point were somehow variable between the two sexes, the directionality of the signature was congruent between males and females at the later time point, with an overall decrease in responses when compared to controls. Interestingly, a group of metabolites collectively known as acylcarnitines and involved in fatty acid b oxidation showed significant responses only in male samples, indicating distinctive biological responses to neutrons between the two sexes implicating mitochondria that should be further explored.
Furthermore, the signatures were able to distinguish the percentage of neutrons in a neutron-photon exposure. This will be of particular importance in a post-IND scenario when appropriate treatment will have to be provided based on the type of radiation exposure. Figure 3 shows that the developed signatures were able to effectively distinguish between the three groups (5, 15 and 25% neutrons in the total dose of 3 Gy), resulting in increased clustering that was more evident with increased time. Once again, for both serum and urine, the results were more prominent in the male cohort. Similar results were previously observed with lipidomic responses (22), with statistically significant lipids exhibiting a noticeable increase at day 7 compared to day 1. Therefore, unlike multiple published studies focusing on gamma or X rays (5, 36–39) showing immediate and significant changes, exposures that contain a percentage of neutrons demonstrate the most prominent results when assessed a few days after irradiation. This is a key finding for logistical reasons during an event. Realistically, samples will be collected at a minimum of 48 h postirradiation, as individuals will still be sheltering in place or awaiting rescue inside collapsed buildings, and therefore arrivals at triage centers will not be immediate. Therefore, a signature that shows an early signal, amplified over time due to underlying biological processes and tissue responses, will be more desirable for development of a biodosimetry method.
As with our previously published neutron metabolomic/lipidomic studies (22, 28), it should be noted that translating these findings to real-life situations involving humans is a difficult endeavor, as animal experiments are tightly controlled and may not accurately represent human physiology. Nonetheless, we were able to show through this study that neutron-photon exposures can be distinguished based on metabolomic signatures and for the first time extended such results with a neutron component to a female cohort. Future studies are needed to validate the results of this work in higher-order primates and human populations.
Supplementary Material
Table S1. Chromatographic and mass spectrometry conditions.
Table S2. Mean and SEM of biomarkers in urine. Statistically significant in relationship to sham.
Table S3. Mean and SEM of biomarkers in serum. Statistically significant in relationship to sham.
Table S4. Statistically significant differences of neutron compared to X-ray-irradiated groups in urine.
Table S5. Statistically significant differences of neutron compared to X-ray irradiated groups in serum.
Fig. S1. sPLS-DA score plots based on the developed signatures. Analysis was conducted between controls (C), X-ray (X) and groups containing a neutron component (N) (pure neutrons, 5%, 15% and 25% grouped as one). Responses were more pronounced with increased time for both male and females.
ACKNOWLEDGMENTS
This work was funded by the National Institutes of Health (NIH; National Institute of Allergy and Infectious Diseases grant no. U19 AI067773; P.I. David J. Brenner, performed as part of Columbia University Center for Medical Countermeasures against Radiation). Dr. Laiakis was also supported by the pilot award AWD-7773347 (P.I. Evagelia C. Laiakis). The project described above was also supported by award no. P30 CA051008 (P.I. Louis Weiner) from the National Cancer Institute (NCI). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the NIH.
Footnotes
Editor’s note. The online version of this article (DOI: https://doi.org/10.1667/RADE-20-00032.1) contains supplementary information that is available to all authorized users.
REFERENCES
- 1.Ainsbury E, Badie C, Barnard S, Manning G, Moquet J, Abend M, et al. Integration of new biological and physical retrospective dosimetry methods into EU emergency response plans – joint RENEB and EURADOS inter-laboratory comparisons. Int J Radiat Biol 2017; 93:99–109. [DOI] [PubMed] [Google Scholar]
- 2.Bensimon Etzol J, Bouvet S, Bettencourt C, Altmeyer S, Paget V, Ugolin N, et al. DosiKit, a new immunoassay for fast radiation biodosimetry of hair and blood samples. Radiat Res 2018; 190:473–82. [DOI] [PubMed] [Google Scholar]
- 3.Coleman CN, Koerner JF. Biodosimetry: Medicine, science, and systems to support the medical decision-maker following a large scale nuclear or radiation incident. Radiat Prot Dosimetry 2016; 172:38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crespo RH, Domene MM, Rodríguez MJ. Biodosimetry and assessment of radiation dose. Rep Pract Oncol Radiother 2011; 16:131–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kultova G, Tichy A, Rehulkova H, Myslivcova-Fucikova A. The hunt for radiation biomarkers: current situation. Int J Radiat Biol 2020; 1–13. [DOI] [PubMed] [Google Scholar]
- 6.Lacombe J, Sima C, Amundson SA, Zenhausern F. Candidate gene biodosimetry markers of exposure to external ionizing radiation in human blood: A systematic review. PLoS One 2018; 13:e0198851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee Y, Pujol Canadell M, Shuryak I, Perrier JR, Taveras M, Patel P, et al. Candidate protein markers for radiation biodosimetry in the hematopoietically humanized mouse model. Sci Rep 2018; 8:13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marchetti F, Coleman MA, Jones IM, Wyrobek AJ. Candidate protein biodosimeters of human exposure to ionizing radiation. Int J Radiat Biol 2006; 82:605–39. [DOI] [PubMed] [Google Scholar]
- 9.Sproull MT, Camphausen KA, Koblentz GD. Biodosimetry: A future tool for medical management of radiological emergencies. Health Secur 2017; 15:599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sullivan JM, Prasanna PG, Grace MB, Wathen LK, Wallace RL, Koerner JF, et al. Assessment of biodosimetry methods for a mass-casualty radiological incident: medical response and management considerations. Health Phys 2013; 105:540–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garty G, Xu Y, Elliston C, Marino SA, Randers-Pehrson G, Brenner DJ. Mice and the A-bomb: Irradiation systems for realistic exposure scenarios. Radiat Res 2017; 187:465–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poeton RW, Glines WM, Mcbaugh D. Planning for the worst in Washington State: initial response planning for improvised nuclear device explosions. Health Phys 2009; 96:19–26. [DOI] [PubMed] [Google Scholar]
- 13.Buddemeier BR. Nuclear detonation fallout: Key considerations for internal exposure and population monitoring. Report no. LLNL-TR-754319 Livermore CA: Lawrence Livermore National Laboratory; 2018. [Google Scholar]
- 14.Cordova KA, Cullings HM. Assessing the relative biological effectiveness of neutrons across organs of varying depth among the atomic bomb survivors. Radiat Res 2019; 192:380–7. [DOI] [PubMed] [Google Scholar]
- 15.Sasaki MS, Endo S, Hoshi M, Nomura T. Neutron relative biological effectiveness in Hiroshima and Nagasaki atomic bomb survivors: a critical review. J Radiat Res 2016; 57:583–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cullings HM, Pierce DA, Kellerer AM. Accounting for neutron exposure in the Japanese atomic bomb survivors. Radiat Res 2014; 182:587–98. [DOI] [PubMed] [Google Scholar]
- 17.Grant EJ, Brenner A, Sugiyama H, Sakata R, Sadakane A, Utada M, et al. Solid cancer incidence among the Life Span Study of Atomic Bomb Survivors: 1958–2009. Radiat Res 2017; 187:513–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Broustas CG, Xu Y, Harken AD, Garty G, Amundson SA. Comparison of gene expression response to neutron and x-ray irradiation using mouse blood. BMC Genomics 2017; 18:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Broustas CG, Xu Y, Harken AD, Chowdhury M, Garty G, Amundson SA. Impact of neutron exposure on global gene expression in a human peripheral blood model. Radiat Res 2017; 187:433–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Broustas CG, Harken AD, Garty G, Amundson SA. Identification of differentially expressed genes and pathways in mice exposed to mixed field neutron/photon radiation. BMC Genomics 2018; 19:504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mukherjee S, Grilj V, Broustas CG, Ghandhi SA, Harken AD, Garty G, et al. Human transcriptomic response to mixed neutron-photon exposures relevant to an improvised nuclear device. Radiat Res 2019; 192:189–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laiakis EC, Canadell MP, Grilj V, Harken AD, Garty GY, Astarita G, et al. Serum lipidomic analysis from mixed neutron/X-ray radiation fields reveals a hyperlipidemic and pro-inflammatory phenotype. Sci Rep 2019; 9:4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Y, Garty G, Marino SA, Massey TN, Randers-Pehrson G, Johnson GW, et al. Novel neutron sources at the Radiological Research Accelerator Facility. J Instrum 2012; 7:C03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Y, Randers-Pehrson G, Turner HC, Marino SA, Geard CR, Brenner DJ, et al. Accelerator-based biological irradiation facility simulating neutron exposure from an improvised nuclear device. Radiat Res 2015; 184:404–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borak TB, Heilbronn LH, Krumland N, Weil MM. Design and dosimetry of a facility to study health effects following exposures to fission neutrons at low dose rates for long durations. Int J Radiat Biol 2019; 1–14. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 26.Cary LH, Ngudiankama BF, Salber RE, Ledney GD, Whitnall MH. Efficacy of radiation countermeasures depends on radiation quality. Radiat Res 2012; 177:663–75. [DOI] [PubMed] [Google Scholar]
- 27.Ledney GD, Elliott TB. Combined injury: factors with potential to impact radiation dose assessments. Health Phys 2010; 98:145–52. [DOI] [PubMed] [Google Scholar]
- 28.Laiakis EC, Wang YW, Young EF, Harken AD, Xu Y, Smilenov L, et al. Metabolic dysregulation after neutron exposures expected from an improvised nuclear device. Radiat Res 2017; 188:21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rossi HH, Bateman JL, Bond VP, Goodman LJ, Stickley EE. The dependence of RBE on the energy of fast neutrons. 1. Physical design and measurement of absorbed dose. Radiat Res 1960; 13:503–20. [PubMed] [Google Scholar]
- 30.Guijas C, Montenegro-Burke JR, Domingo-Almenara X, Palermo A, Warth B, Hermann G, et al. METLIN: A technology platform for identifying knowns and unknowns. Anal Chem 2018; 90:3156–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fahy E, Cotter D, Sud M, Subramaniam S. Lipid classification, structures and tools. Biochim Biophys Acta 2011; 1811:637–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vazquez-Fresno R, et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res 2018; 46:D608–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res 2018; 46:W486–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Cao KA, Boitard S, Besse P. Sparse PLS discriminant analysis: biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinformatics 2011; 12:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kramer K, Li A, Madrigal J, Sanchez B, Millage K. Monte-Carlo modeling of the prompt radiation emitted by a nuclear device in the national capital region, revision 1. Report No. DTRA-TR-13–045 (R1) Fort Belvoir, VA: Defense Threat Reduction Agency; 2016. [Google Scholar]
- 36.Sproull M, Camphausen K. State-of-the-art advances in radiation biodosimetry for mass casualty events involving radiation exposure. Radiat Res 2016; 186:423–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coy SL, Cheema AK, Tyburski JB, Laiakis EC, Collins SP, Fornace A Jr. Radiation metabolomics and its potential in biodosimetry. Int J Radiat Biol 2011; 87:802–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Menon SS, Uppal M, Randhawa S, Cheema MS, Aghdam N, Usala RL, et al. Radiation metabolomics: current status and future directions. Front Oncol 2016; 6:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pannkuk EL, Fornace AJ Jr., Laiakis EC. Metabolomic applications in radiation biodosimetry: exploring radiation effects through small molecules. Int J Radiat Biol 2017; 93:1151–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Chromatographic and mass spectrometry conditions.
Table S2. Mean and SEM of biomarkers in urine. Statistically significant in relationship to sham.
Table S3. Mean and SEM of biomarkers in serum. Statistically significant in relationship to sham.
Table S4. Statistically significant differences of neutron compared to X-ray-irradiated groups in urine.
Table S5. Statistically significant differences of neutron compared to X-ray irradiated groups in serum.
Fig. S1. sPLS-DA score plots based on the developed signatures. Analysis was conducted between controls (C), X-ray (X) and groups containing a neutron component (N) (pure neutrons, 5%, 15% and 25% grouped as one). Responses were more pronounced with increased time for both male and females.


