Figure 1. Differential labeling of ribosomes reveals variation of ribosome biogenesis and turnover across different cell types and conditions.
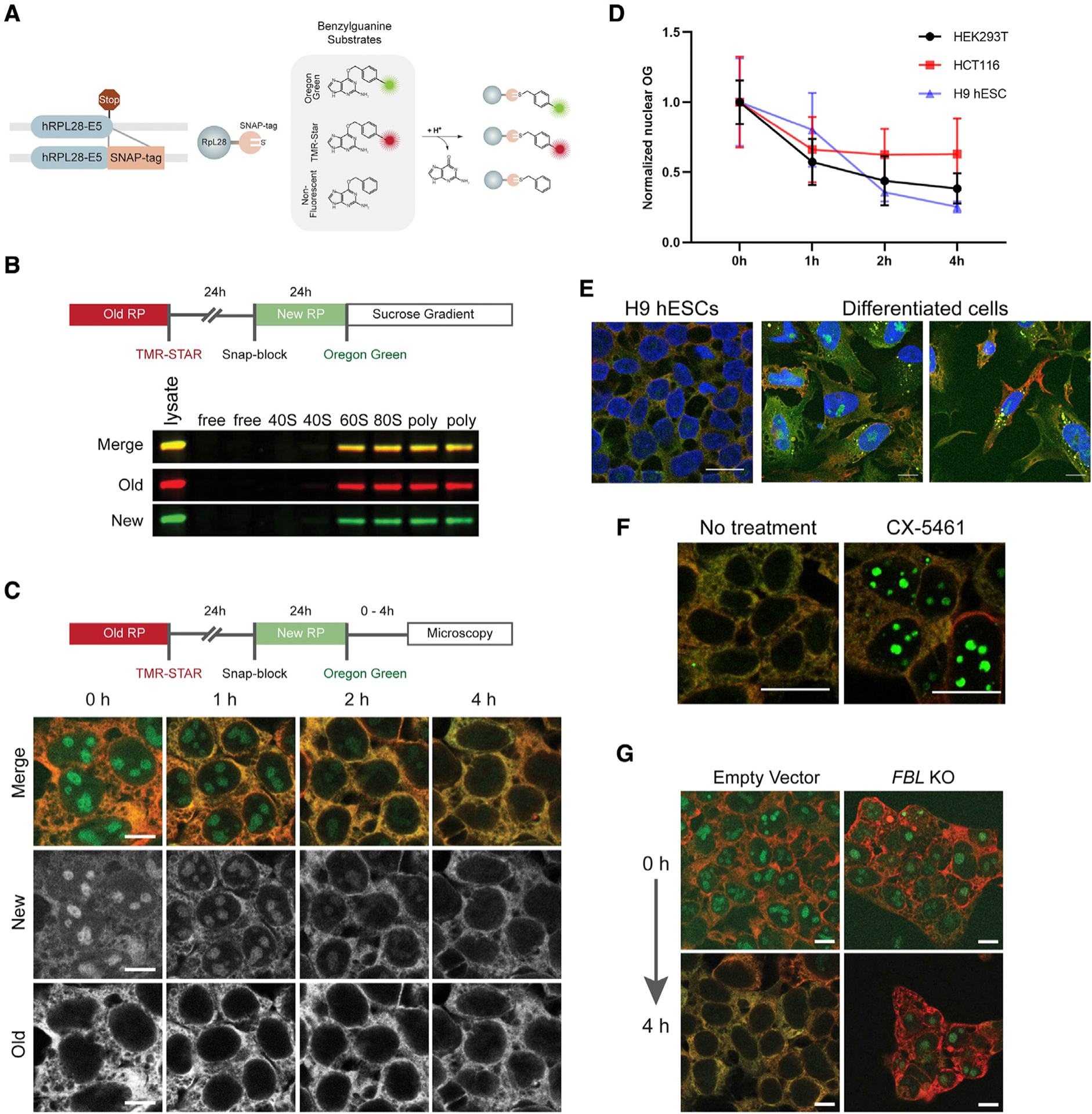
(A) Schematic detailing the tagging of the RPL28 open reading frame with a SNAP tag at the 3′ end of exon 5 (E5). The SNAP-tagged proteins can be indelibly labeled with a number of different benzylguanine substrates. They can also be “blocked” through the use of unconjugated benzylguanine.
(B) An outline of a standard pulse-labeling experiment in which ribosomes older than 48 h are labeled with TMR-Star (red) and ribosomes 0–24 h old are labeled with Oregon green. Cell lysates were subjected to sucrose gradient fractionation. Old and newly labeled RPL28 incorporated into the 60S, monosome, and polysome fractions.
(C) Imaging of pulse-labeled Ribo-SNAP cells. After the final labeling step, the cells are incubated for variable lengths of time. At the start of the experiment, newly labeled RPL28 is clearly observed in the nucleoli of cells, whereas old ribosomes are cytoplasmic, as expected. The nucleolar levels of newly labeled RPL28 continue to drop over the course of 4 h.
(D) The rate at which the levels of nuclear RPL28 decreases varies between HEK293T, HCT116, and hESCs. Values were normalized to the mean of the 0-h group of each cell type. Mean ± SD is shown.
(E) H9 hESCs carrying the RPL28 SNAP tag reporter or their randomly differentiated progeny were pulse labeled for old and new ribosomes and examined after a 4-h chase. Levels of labeled ribosomes appear relatively homogeneous within hESCs but their differentiated progeny displayed a range of old and new ribosome levels. Moreover, differentiated cells exhibited variable amounts of newly labeled nucleolar RPL28 after a 4-h chase period.
(F) Treatment of cells with the Pol I and TOP2B inhibitor CX-5461 results in retention of newly labeled RPL28 within nucleoli after a 4-h chase period.
(G) Cells carrying SNAP-tagged RPL28 were transduced with sgRNAs targeting FBL and subjected to pulse labeling. Disruption of FBL resulted in retention of newly labeled RPL28 within nucleoli after a 4-h chase period. Scale bars, 10 μm in (C, E, and G) and 20 μm in (F).
