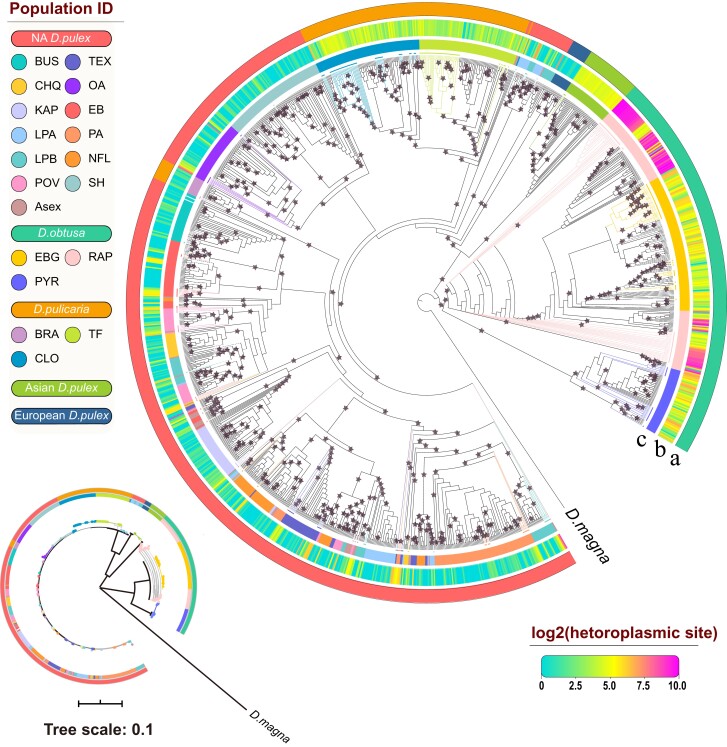
Fig. 3.
Mitochondrial phylogeny of the D. pulex, D. pulicaria, and D. obtusa clones based on maximum-likelihood analysis of the full-length mitochondrial sequences. Daphnia magna was used as an outgroup. Clones with two haplotypes are phased using allele frequencies, that is, assigning all major alleles to one haplotype, and all minor alleles to the other. Haplotypes constructed from minor alleles are marked by colored branches and solid circles at the tip of the corresponding branches. Arc (a) shows the color-coded species, with species name listed on the left panel; Arc (b) indicates the density of the heteroplasmic sites within each clone; Arc (c) shows the color-coded populations within each species. Stars indicate bootstrap values >75%. Inset on the bottom left shows the branch length of the major clades.