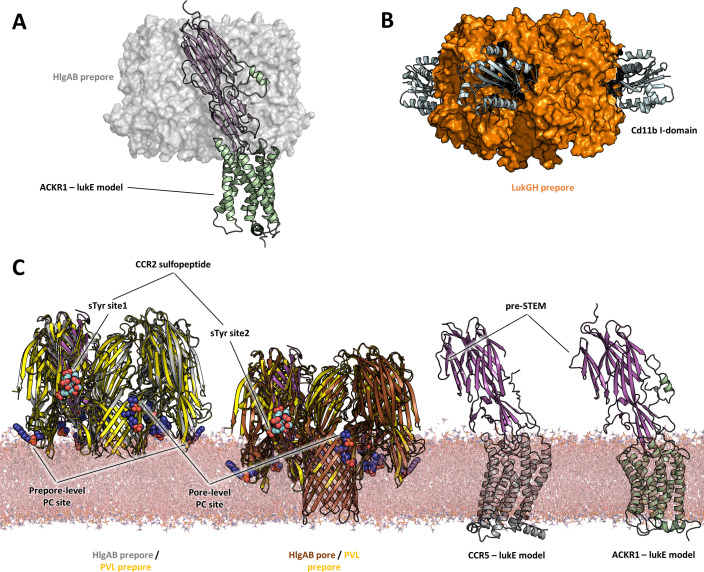
Figure 10. Comparisons of LukE-CCR2 peptide x-ray structure, ACKR1-LukE and CCR5-LukE models with existing x-ray structures.
(A) Structural alignment of LukE from the ACKR1-LukE model with the HlgAB prepore structure (PDB ID 4P1Y), illustrating that the N-terminal region of ACKR1 binds to a surface located on the outer part of the prepore. The HlgAB prepore is shown as a semitransparent surface and the ACKR1-LukE model as cartoon. (C) X-ray structure of LukGH bound to human CD11b I-domain. The LukGH prepore is shown as a semitransparent surface and the CD11b I-domain as cartoon. (C) The HlgAB pore and prepore structures (PDB IDs 3B07 and 4P1Y) are shown in their membrane context using a POPC bilayer extracted from MDS simulations. On the left, the HlgAB pore and prepore structures are superimposed to the PVL prepore (PDB ID 6U3Y) bound with fos-choline-14 (shown as blue spheres). One of the lukF-PV/HlgB protomer is omitted for clarity. The structural alignment highlights the presence of lipid headgroup binding sites on lukF-PV that correspond to different membrane insertion depths, which we interpreted as pore and prepore membrane levels. The LukE-CCR2 peptide structure is also superimposed onto the pore and prepore structures to indicate the location of the bound sulfotyrosines (shown as cyan spheres). In the case of a fully formed pore, sTyr sulfate at site 2 is located at the same level as the PC headgroups. On the right, the ACKR1-LukE and CCR5-LukE models are shown in their membrane context, and oriented to visualize the acute angle between LukE and the membrane.