Abstract
Since the COVID-19 pandemic, several research studies have proposed Deep Learning (DL)-based automated COVID-19 detection, reporting high cross-validation accuracy when classifying COVID-19 patients from normal or other common Pneumonia. Although the reported outcomes are very high in most cases, these results were obtained without an independent test set from a separate data source(s). DL models are likely to overfit training data distribution when independent test sets are not utilized or are prone to learn dataset-specific artifacts rather than the actual disease characteristics and underlying pathology. This study aims to assess the promise of such DL methods and datasets by investigating the key challenges and issues by examining the compositions of the available public image datasets and designing different experimental setups. A convolutional neural network-based network, called CVR-Net (COVID-19 Recognition Network), has been proposed for conducting comprehensive experiments to validate our hypothesis. The presented end-to-end CVR-Net is a multi-scale-multi-encoder ensemble model that aggregates the outputs from two different encoders and their different scales to convey the final prediction probability. Three different classification tasks, such as 2-, 3-, 4-classes, are designed where the train–test datasets are from the single, multiple, and independent sources. The obtained binary classification accuracy is 99.8% for a single train–test data source, where the accuracies fall to 98.4% and 88.7% when multiple and independent train–test data sources are utilized. Similar outcomes are noticed in multi-class categorization tasks for single, multiple, and independent data sources, highlighting the challenges in developing DL models with the existing public datasets without an independent test set from a separate dataset. Such a result concludes a requirement for a better-designed dataset for developing DL tools applicable in actual clinical settings. The dataset should have an independent test set; for a single machine or hospital source, have a more balanced set of images for all the prediction classes; and have a balanced dataset from several hospitals and demography. Our source codes and model are publicly available1 for the research community for further improvements.
Keywords: COVID-19 disease, Chest computed tomography and X-ray, Convolutional neural networks, Ensemble classifier
1. Introduction
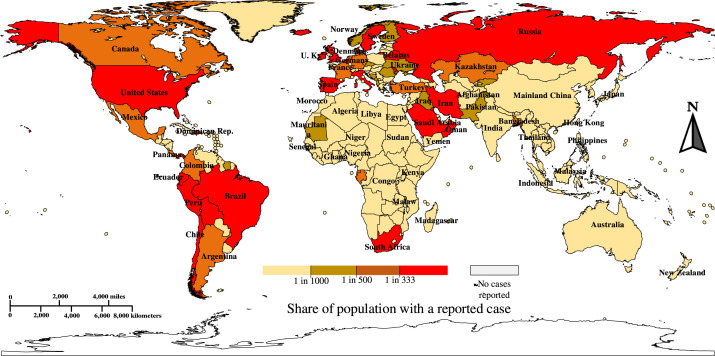
Pneumonia of unknown cause detected in Wuhan, China, was reported to the World Health Organization (WHO) office in China on 31st December 2019. This was subsequently named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) on 11th February 2020, as the virus causing the disease is genetically related to the coronavirus responsible for the SARS outbreak of 2003. The new disease was referred to as “COVID-19” by WHO on 11th February 2020 [1]. As of August , the outbreak of in Wuhan (China), has extended worldwide with confirmed COVID- cases including deaths in last 2 years (5 February 2022) [2], as presented in Fig. 1. The clinical attributes of severe COVID- epidemic are bronchopneumonia that causes cough, fever, dyspnea, and subtle respiratory anxiety ailment [3], [4], [5]. The clinical screening test for COVID- is Reverse Transcription Polymerase Chain Reaction (RT-PCR) using respiratory specimens. However, this test is a manual, complicated, tedious, and time-consuming procedure with an estimated true-positive rate of 63.0% [6]. There is a significant lack of inventory of RT-PCR kits, leading to a delay in efforts to prevent and cure coronavirus disease [7]. Furthermore, the RT-PCR kit is estimated to cost around USD and requires a specially designed biosafety laboratory to house the PCR unit, each of which can cost USD [8]. Nevertheless, the utilization of a costly screening device with delayed test results makes it more challenging to suppress the spread of the disease.
Fig. 1.
A world heat map of the corona pandemic per capita [9] [Accessed on 25 December 2021].
However, it is observed that most of the COVID- cases have common characteristics on radiographic images, such as Computed Tomography (CT) and Chest X-ray (CXR), including bilateral, multi-focal, ground-glass opacities with a peripheral or posterior distribution, mainly in the lower lobes and early- and late-stage pulmonary consolidation [10], [11], [12], [13]. Those features can be utilized to develop a sensitive Computer-aided Diagnosis (CAD) tool to detect COVID- Pneumonia and be considered as a screening tool [14]. Currently, deep Convolutional Neural Networks (CNNs) allow for building an end-to-end model, without the need for manual feature extraction [15], [16], which have demonstrated tremendous success in many domains of medical imaging, such as arrhythmia detection [17], [18], [19], skin lesion segmentation and classification [20], [21], [22], [23], [24], breast cancer detection [25], [26], [27], brain disease classification [28], pneumonia detection from CXR images [29], fundus image segmentation [30], [31], minimally invasive surgery [32] and lung segmentation [33]. Several deep CNN-based methods have been published to detect COVID-19 from CXR and CT images. Though the results obtained are promising, they exhibit limited scope as a CAD tool. Most of the works, especially on CXR images, have been based on data from different sources for two different classes (COVID vs. Normal). This brings inherent bias on the algorithms as the model tends to learn the distribution and artifacts of the data source for binary classification problems. Therefore, these models perform very poorly when used in practical settings where the model has to adapt to data from different domains. To accelerate the development of DL tools that could be utilized in realistic clinical settings, the scientific community needs to emphasize more on making publicly systematically-designed and documented datasets that have information, such as inclusion and exclusion criteria, symptomatic vs. asymptomatic cases, and the disease severity stage at which these images were taken. In this work, we design various experiments with a proposed CNN-based COVID-19 detection method to justify this proposition.
The rest of the paper is structured as follows: Section 2 reviews the earlier published literature for COVID-19 detection, and Section 3 highlights the significant contributions to this article. We explain the proposed framework for the recognition of COVID-19 and datasets in Section 4. The results and different experiments are reported in Section 5. We interpret the obtained results from the proposed CVR-Net in Section 6. Finally, Section 7 concludes the article with future working directions.
2. Review of literature
Different CNN architectures have already been proposed for COVID-19 detection as a binary (COVID vs. No-Findings) or multi-class (COVID vs. No-Findings vs. Pneumonia) problem [34], [35], [36]. Ghoshal and Tucker [37] investigated uncertainty of the COVID-19 classification report, using a drop-weights-based Bayesian CNN, as the availability of uncertainty-aware DL can ensure more extensive adoption of DL in clinical applications. Abbas et al. [38] proposed a framework by adopting a deep CNN, called Decompose, Transfer, and Compose (DeTraC) [39] for the classification of COVID- CXR images, where the authors implemented the DeTraC in two phases. Firstly, using gradient descent optimization, they trained the backbone pre-trained CNN model of DeTraC to extract deep local features from each image. Secondly, they used the class-composition layer of DeTraC to refine the final classification of the images. Zhao et al. [40] developed diagnosis methods based on multi-task learning and self-supervised learning, where the authors proposed an open-source COVID-19 dataset of CT images with a binary class (COVID and Non-COVID). For the classification task, they trained DenseNet- and ResNet-, via a pre-trained model on ImageNet [16] weights, with their newly proposed dataset. Afshar et al. [41] proposed a CNN model named COVID-CAPS, which was based on the Capsule Networks (CapsNets) for handling the small datasets of COVID-19. CapsNets are alternative models of CNN, which are capable of capturing spatial information using routing by agreement. Capsules try to reach a mutual agreement on the existence of the objects. Their proposed COVID-CAPS model had convolutional layers and capsule layers, where batch normalization [42] followed the former layers. The authors fine-tuned all the capsule layers, while the conventional layers were frozen with pre-trained weights of ImageNet. He et al. [43] built a COVID-19 CT dataset, called China Consortium of Chest CT Image Investigation (CC-CCII), with three classes: novel coronavirus Pneumonia, common Pneumonia, and healthy controls. The authors trained D DenseNet3D- on their proposed CC-CCII dataset, and they experimentally validated that D CNNs outperform D CNNs in general. Singh et al. [13] implemented a CNN-based model named multi-objective differential evolution-based CNN for the classification of COVID-. They fine-tuned the parameters of the CNN model using a multi-objective fitness function. The differential evolution algorithm was used to optimize the multi-objective fitness function. The model was optimized iteratively using mutation, crossover, and selection operation to determine the best available solution in differential evolution. Farooq and Hafeez [44] employed ResNet- using transfer learning with progressively resizing [45] the input images to 128 × 128 × 3, 224 × 224 × 3, and 229 × 229 × 3 pixels, where the authors also fine-tuned the network at each stage. Ozkaya et al. [46] extracted deep features using VGG-, GoogleNet [47], and ResNet- models, which were classified by Support Vector Machine (SVM) [48] with linear kernel function. They also applied the modified T-test [49], a feature ranking algorithm, to select the features [50] for avoiding overfitting. Rajaraman et al. [51] evaluated ImageNet pre-trained CNN models such as VGG-, VGG-, InceptionV, Xception, Inception-ResNetV, MobileNetV, DenseNet-, and NasNet-mobile [52]. Then, they optimized the hyperparameters of the CNNs using a randomized grid search method [53]. In the end, the authors proposed an ensemble of those CNN models for the final COVID-19 recognition. Toğaçar et al. [54] restructured the data classes using a fuzzy color technique, where they stacked a structured image with the original images. The authors trained MobileNetV and SqueezeNet to extract the deep features, which were then processed using the social mimic optimization method [55]. After that, selected features were combined and classified using the SVM to recognize COVID-19. Khan et al. [56] developed a 15-layered CNN architecture for extracting deep features from two different layers like global average pool and fully connected layers, which were then merged employing the max-layer detail approach. The most discriminant features from the pool of features were selected using a Correntropy technique, and a one-class kernel extreme learning machine classifier was applied for the classification. CNN-based models like ResNet50, ResNet101, ResNet152, InceptionV3, and Inception-ResNetV2 were proposed and implemented by Narin et al. [57] for the detection of COVID-19-infected patient using CXR radiographs. Sedik et al. [58] classified CT and CXR images of COVID-19 vs. normal using CNN and convolutional long short-term memory (ConvLSTM) based models. Sanida et al. [59] employed lightweight modified MobileNetV2 to classify the COVID-19, normal, viral Pneumonia, and lung opacity images for the real-time operations in a low-power embedded system. Authors in [60] proposed a COVIDetectioNet using a pre-trained AlexNet to extract the deep features. The useful features were selected using the Relief algorithm from all layers of the architecture were then classified using the SVM approach. An efficient Grayscale Spatial Exploitation Net (GSEN) is designed by employing web pages crawling across cloud computing environments in [61], utilizing the accuracy rates improvement in a positive relationship to the cardinality of crawled CXR dataset. Their model consists of four convolutional blocks where each is composed of a single convolutional, batch normalization, ReLU activation function, and max-pooling layer. Monday et al. [62] proposed a neurowavelet capsule network. Firstly, they presented a multi-resolution analysis of a discrete wavelet transform to filter noisy and incompatible information from the CXR data to enhance the feature extraction robustness of the network. Secondly, the discrete wavelet transform of the multi-resolution analysis was also conducted a sub-sampling procedure to minimize the loss of spatial details, thereby improving the overall classification performance. Sakthivel et al. [63] proposed an ensemble-based CNN model where five DL models like ResNet, FitNet, IRCNN, MobileNet, and EfficientNet are ensembled and fine-tuned to classify the CXR images. An application-specific hardware architecture had been incorporated by carefully exploiting the data flow and resource availability.
3. Our contributions
Many DL-based Artificial Intelligence (AI) algorithms have been proposed in the past year to automatically classify COVID-19 cases from normal and other Pneumonia cases. These published works reported high COVID-19 binary classification accuracy using either CT scans or CXRs [13], [34], [35], [45], [54], [64], [65], [66], [67], [68], [69], [70], [71]. Although the reported metrics, such as sensitivity and/or specificity, are very high in most cases, these results are obtained on cross-validation studies without an independent test set coming from a separate dataset having biases, such as the two classes predicted from two unique datasets. AI models are likely to overfit training data distribution when independent test sets are not used or are prone to learn dataset-specific artifacts rather than the actual disease characteristics. Additionally, the publicly available datasets for COVID-19 classification used in the recent studies have class and dataset source biases, resulting in AI models learning dataset-specific distributions rather than the underlying pathology. Many recent studies proposing COVID-19 classification based on DL using imaging data do not emphasize the importance of avoiding overfitting and having an independent test set with images from a separate dataset than the training and validation dataset. However, the critical contributions in this article are pointed out as follows:
-
•
Proposing an end-to-end and multi-scale-multi-encoder CVR-Net, aggregating the outputs from two different encoders and their different scales to obtain the final prediction probability.
-
•
Designing various experiments to investigate the issues of overfitting and biasing; exploring the limitations of existing large public datasets that have been widely used for developing and evaluating COVID-19 detection algorithms in the past years.
-
•
Validating multi-class classification models to distinguish various Pneumonia types, including COVID-19, requires a balanced set of images for all the prediction classes coming from a single site and demography and having several balanced sets coming from separate scanners or hospitals and demography.
-
•
Comparing the proposed architecture to other state-of-the-art methods using an independent test set for evaluation, where some of the identified bias and overfitting issues are minimized.
4. Materials and methods
This section presents the materials and methods for conducting this research. Section 4.1 briefly describes utilized datasets. The designing of the proposed network (CVR-Net) is explained in Section 4.2. Finally, Section 4.3 describes the training protocol of our network and the evaluation metrics.
4.1. Datasets
This section illustrates the experimental setup for various classification tasks utilizing chest CT scans or CXRs from several publicly available datasets. The classes applied for different experimentations are taken from the following set:
-
•
NOR: Normal; no Pneumonia and COVID-19 negative
-
•
CVP: COVID-19 positive Pneumonia
-
•
OVP: Other Viral Pneumonia; Viral Pneumonia but not COVID-19
-
•
OBP: Other Bacterial Pneumonia; Bacteria induced Non-COVID Pneumonia
-
•
NCP: Non-COVID Pneumonia; OBP + OVP
-
•
NCV: Non-COVID; NOR + NCP
Table 1 demonstrates the details of the experimental setup with various tasks and how various datasets are combined for these tasks.
Table 1.
Various classification tasks utilizing CT scans or CXRs in different combinations from publicly available datasets.
| Different studiesa | Class categories | # of images | Data source references | Modality | Utilization |
|---|---|---|---|---|---|
| CXR-Single-CL2 | NCV | 5,856 | CXRI [72] | X-ray | [34], [35], [65] |
| CVP | 500 | CIDC [73] | |||
| CXR-Multiple-CL2 | NCV | 7,864 | CXRIL [72], ChestX-ray8 [74] | X-ray | Proposed |
| CVP | 4,015 | CCXRIL [75], CIDC [73], PadChest [76] | |||
| CXR-Independent-CL2 | NCV (TrainTest) | CheXpert [77]+ CXRI [72]ChestX-ray8 [74] | X-ray | Proposed | |
| CVP (TrainTest) | CCXRI [75]+ PadChest [76]CIDC [73] | ||||
| CT-Single-CL2 | NCV | 1,227 | SCoV [78] | CT | Proposed |
| CVP | 1,252 | SCoV [78] | |||
| CT-Multiple-CL2 | NCV | 7,864 | SCoVL [78], CCII [71], MGC [40] | ||
| CVP | 4,015 | SCoVL [78], CCII [71], MGC [40] | CT | Proposed | |
| CT-Independent-CL2 | NCV (TrainTest) | MGC [40]+ CCII [71]+ iCTCF [79]SCoV [78] | CT | Proposed | |
| CVP (TrainTest) | MGC[40]+ CCII [71]+ iCTCF [79]SCoV [78] | ||||
| CXR-Single-CL3 | NOR | 1,583 | CXRI [72] | X-ray |
[65], [80] [34], [35] |
| NCP | 4,273 | CXRI [72] | |||
| CVP | 500 | CIDC [73] | |||
| CXR-Multiple-CL3 | NOR | 3,591 | CXRIL [72], ChestX-ray8 [74] | X-ray | Proposed |
| NCP | 4,595 | CXRIL [72], ChestX-ray8 [74] | |||
| CVP | 4,015 | CCXRIL [75], CIDC [73], PadChest [76] | |||
| CXR-Multiple-CL4 | NOR | 3,591 | CXRIL [72], ChestX-ray8 [74] | X-ray | Proposed |
| OBP | 2,780 | CXRI [72] | |||
| OVP | 1,493 | CXRI [72] | |||
| CVP | 4,015 | CCXRIL [75], CIDC [73], PadChest [76] | |||
X-Y-CL#: X is CXR or CT; Y denotes the way images from different sources are combined for each class during training or evaluation; CL# is the number of classes.
Three different types of classification tasks are designed: NCV vs. CVP (-classes, CL2); NOR vs. NCP vs. CVP (-classes, CL3); and NOR vs. OBP vs. OVP vs. CVP (-classes, CL4). Several different combinations of the publicly available datasets are utilized for chest CT scans (labeled CT) and for chest X-rays (labeled CXR) [40], [71], [72], [73], [74], [75], [76], [77], [78], [79]. For each binary (CL2) or multi-class (CL3/CL4) classification task, we design experiments to study the impact of having single separate vs. multiple mixed sources of data for individual classes during training, labeled Single and Multiple, respectively. The setup where the test set contains images from an independent source whose images are never used during training and validation is labeled as Independent. For adding diversity in each class of CXR-Multiple-CL2, we include images from more datasets: CXR images from ChestX-ray8 [74] to NCV, and from CCXRI [75] and PadChest [76] to CVP. To evaluate the ability to distinguish various Pneumonia types, we design CXR-Multiple-CL3 and CXR-Multiple-CL4 having the same number of images as CXR-Multiple-CL2, but the NCV class split into individual Pneumonia types.
Similar to CXR, publicly available CT scan datasets are also utilized, where most of these datasets contained manually selected 2D slices instead of complete 3D volumes. Hence, all of the CT images referred to in this paper are 2D slices of CT scans. CT-Single-CL2 utilizes NCV and CVP samples from SCoV [78], while we have multiple sources to each class in CT-Multiple-CL2 adding NCV and CVP samples from MGC [40], SCoV [78], and CCII [71]. Due to a lack of publicly available images, some of the designs were not possible, for example, CT-Multiple-CL3 and CT-Multiple-CL4. To evaluate the network’s performance on an independent test set from a separate dataset source whose images are never used during the network’s training, we design CXR-Independent-CL2 and CT-Independent-CL2, utilizing train data from a large study in Spain and test data from the other sources. Table 1 details the train/test split for these two setups. Fig. 2 shows example images from these datasets. In the setup where an independent test dataset is not available, 5-fold cross-validation is applied to evaluate the performance of the proposed CVR-Net (see in Section 4.2).
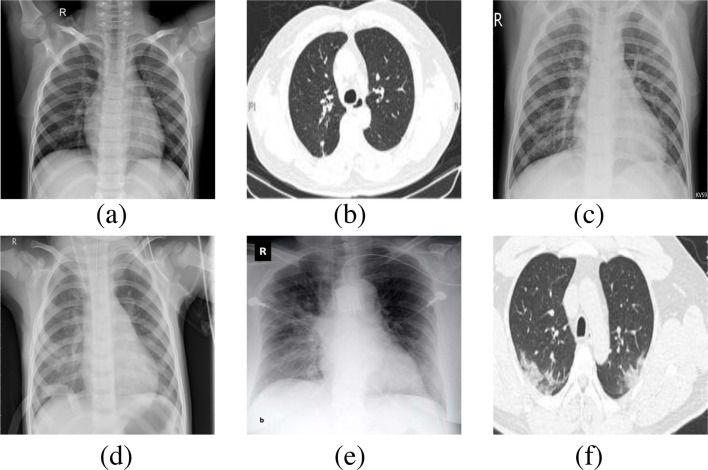
Fig. 2.
Samples of chest radiography images from the utilized datasets (a) Normal (X-ray), (b) Normal (CT), (c) Pneumonia viral (X-ray), (d) Pneumonia bacterial (X-ray), (e) COVID-19 (X-ray), and (f) COVID-19 (CT).
4.2. Proposed CVR-Net Architecture
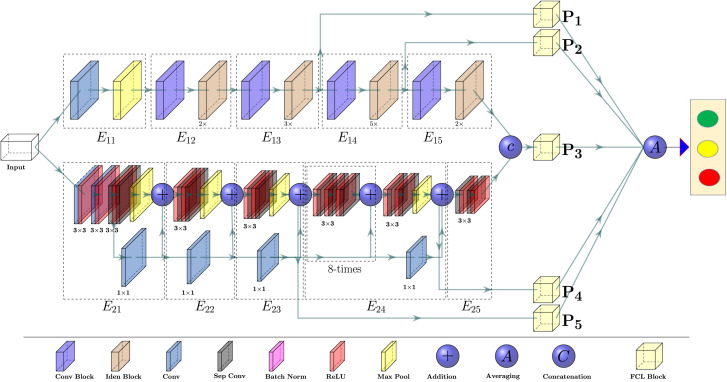
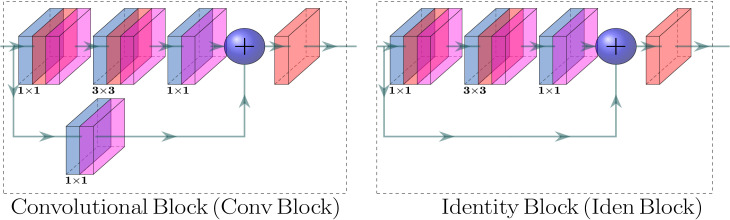
We propose a CNN-based end-to-end multi-tasking network, where we apply multi-encoder and multi-scale ensembling, as depicted in Fig. 3. The proposed CVR-Net consists of two encoders, for the same input image, where each of the encoders has five blocks, namely and , , for encoder- and encoder-, respectively. The encoder-1 consists of the residual and convolutional blocks [81], as presented in Fig. 4, well-known as ResNet [81]. The residual connections, also known as skip connections, allow gradients to flow through a network directly, without passing through non-linear activation functions and thus avoiding the problem of vanishing gradients [81]. In residual connections, the output of a weight layer series is added to the original input and then passed through the non-linear activation function, as shown in Fig. 4. However, in encoder-, 7 × 7 input convolution, followed by max-pooling with the stride of , and pool size of 3 × 3, is used before identity and convolutional blocks. By stacking these blocks on top of each other (see Fig. 3), an encoder- is formed to get the feature map, where the notation () under the identity block denotes the number of repetitions ( times). The different blocks of encoder- ( and ) downsample the input image resolutions in half of the input resolutions, while the resolution inside the blocks is kept constant. The outputs of those blocks generate the feature maps with different scales. Within the encoder- (Xception), three components of information flow blocks are used, which were initially proposed by Chollet [82], such as entry flow, middle flow, and exit flow, as depicted in Fig. 3. The batch of input images first passes through the input flow, then the central flow, eight times () repeated, and finally through the exit flow. All flows, as in the proposed network (see in Fig. 3), have Depth-wise Separable Convolution (DwSC) [82] and residual connections. As in the case of encoder-, the resolution after each block is downsampled by the factor of two, and the exact resolution is maintained at each block for encoder-. After the two encoder blocks, the two different D feature maps are concatenated channel-wise to enhance the depth information of the feature map. We use differently scaled feature maps to build the proposed CVR-Net, where each feature map is passed through the Fully Connected Layer (FCL) block. A Global Average Pooling (GAP) [83] layer and four fully connected layers are used in our FCL block, where the GAP layer performs an extreme dimensionality reduction to avoid overfitting. In GAP, an dimensional tensor is reduced to a vector by transferring feature map to a single number contributes to the lightweight design of the proposed CVR-Net. Table 2 presents the implementational details of the proposed CVR-Net. We utilize the feature maps from encoder- and from encoder-, where we concatenate and to increase the depth of the feature information. The final prediction, in CVR-Net, is the average of different probabilities, such as , , , , and respectively for , , , , and , which was trained end-to-end fashion. However, designing of such a multi-encoder and multi-scale network, as CVR-Net, has several benefits, especially for the small datasets, such as: if one encoder fails to generate responsible features, another encoder can compensate it and vice-versa; if the feature quality is reduced in the deeper blocks (lower resolution), the prior blocks (higher resolution) can also compensate it and vice-versa; if one or more predicts wrong class, other can overcome it, as the final result is average of all ’s. Another positive prospect of the CVR-Net is that during the training, it can be anticipated that if the gradient of one or more branches vanishes, other branches can recover it as the final gradient is the average of all the individual gradients.
Fig. 3.
The proposed network, called CVR-Net, for the automatic COVID-19 recognition from radiography images, where we ensemble the multi-encoder and multi-scale of the network, via fully connected blocks, obtain final recognition probability.
Fig. 4.
The convolutional (left) and residual (right) blocks [81] of the proposed CVR-Net, where the output map is the summation of the input map and the generated map from the process (convolutions).
Table 2.
Details of the proposed CVR-Net have used feature maps, shapes, and the number of parameters, where the input resolution is pixels.
| Feature block | Shape of features | Prediction | Parameters |
|---|---|---|---|
| Proposed CVR-Net | |||
4.3. Training protocol and evaluation
Since most images in all the datasets have a aspect ratio, we resize the images to 224 × 224 pixels using nearest-neighbor interpolation. We apply the following stochastic augmentation on the resized images with: rotation (with a probability of 0.45), height & width shift (with a probability of 0.20), and vertical & horizontal flipping around the X- and -axis (with a probability of 1.0), respectively. We employ categorical cross-entropy as a loss function [84], penalizing the majority class by giving higher weight in the loss function to the samples from the minority class. Each class’s weights are computed as , where and are the weights, and the total number of samples for the th class and is the total sample numbers. The network weights are initialized using transfer learning [85] where we use the ImageNet pre-trained weights of ResNet-50 and Xception to initialize the weights of the two respective branches. We use Adam optimizer to optimize the training network with initial learning rate (), exponential decay rates () as , , and , respectively, without AMSGrad variant [86]. The initial learning rate is reduced after epochs by 10.0% if validation loss stops improving. The training is terminated after epochs if the validation performance stops improving.
The models were implemented using the Python programming language and Keras framework [87] and the experiments were carried out on a machine running Windows-10 operating system with the following hardware configuration: Intel® CoreTM i HQ CPU @ processor with Install memory (RAM): and GeForce GTX GPU with memory. When comparing against other state-of-the-art methods (see in Table 5), the same above-described protocol was operated for all the networks.
Table 5.
Comparison of various methods, including the proposed network (CVR-Net), where the methods are trained on the same dataset and evaluated using an independent test set, not used during training. The top three performing metrics are denoted by bold-font, underline, and double-underline.
| Methods | Parameters | CXR-Independent-CL2 |
CT-Independent-CL2 |
||||
|---|---|---|---|---|---|---|---|
| Recall | Precision | Accuracy | Recall | Precision | Accuracy | ||
| VGG-19 | M | 0.833 | 0.846 | 0.833 | 0.785 | 0.816 | 0.785 |
| Xception | M | 0.869 | 0.881 | 0.869 | 0.718 | 0.788 | 0.718 |
| EfficientNet-b1 | M | 0.832 | 0.850 | 0.832 | 0.716 | 0.803 | 0.716 |
| DenseNet-169 | M | 0.850 | 0.865 | 0.850 | 0.718 | 0.794 | 0.718 |
| ResNet-152 | M | 0.829 | 0.866 | 0.829 | 0.705 | 0.784 | 0.705 |
| Inception-v3 | M | 0.871 | 0.884 | 0.871 | 0.737 | 0.782 | 0.737 |
| DarkNeta[34] | M | 0.712 | 0.699 | 0.712 | 0.495 | 0.245 | 0.495 |
| CoroNeta[35] | M | 0.869 | 0.877 | 0.869 | 0.689 | 0.776 | 0.689 |
| Proposed CVR-Net | M | ||||||
We have implemented those models in our experimental settings for ablation studies.
We use different metrics, such as recall, precision, F1-score, and accuracy, to evaluate our multi-tasking CVR-Net for COVID-19 recognition, which is mathematically defined [88] as follows:
where the TP, FN, FP, and TN respectively denote true positive (patient with coronavirus symptoms recognized as the positive patient), false negative (patient with coronavirus symptoms recognized as the negative patient), false positive (patient without coronavirus symptoms recognized as the positive patient), and true negative (patient without coronavirus symptoms recognized as the negative patient). The recall quantifies the type-II error (the patient, with the positive syndromes, inappropriately fails to be nullified), and precision quantifies the positive predictive values (percentage of truly positive recognition among all the positive recognition). The F-score indicates the harmonic mean of recall and precision, which shows the tradeoff between them. Accuracy quantifies the fraction of correct predictions (both positive and negative).
5. Experimental results
This section initially presents the results of binary and multi-class classification tasks for various setups described in Section 4.1 using the architecture proposed in Section 4.2. Finally, we compare the proposed network’s performance with state-of-the-art classification networks by training them on the same training set and evaluating an independent test set whose images are not used during training.
5.1. Binary classification: COVID vs. Non-COVID
Table 3 presents the quantitative results of the proposed CVR-Net on the binary task: COVID-19 (CVP) vs. Non-COVID (NCV). The 5-fold cross-validation results are conveyed with average and standard deviation. In contrast, a single value is reported when a separate test set from an independent data source is used to evaluate the results.
Table 3.
COVID-19 recognition results from different studies of binary classification applying the proposed network on two different modalities of chest radiography images, wherein for single and multiple sources, we employ 5-fold cross-validation.
| Different studiesa | Dataset distribution | Metrics |
||
|---|---|---|---|---|
| (TrainValTest) | Recall | Precision | Accuracy | |
| NCV: | ||||
| CXR-Single-CL2 | CVP: | |||
| NCV: | ||||
| CXR-Multiple-CL2 | CVP: | |||
| NCV: | ||||
| CXR-Independent-CL2 | CVP: | 0.887 | 0.885 | 0.887 |
| NCV: | ||||
| CT-Single-CL2 | CVP: | |||
| NCV: | ||||
| CT-Multiple-CL2 | CVP: | |||
| NCV: | ||||
| CT-Independent-CL2 | CVP: | 0.799 | 0.821 | 0.799 |
X-Y-CL#: X is CXR or CT; Y denotes the way images from different sources are combined for each class during training or evaluation; CL# is the number of classes. Details in Table 1.
Table 3 demonstrates very high precision and recall in both the cases of CXR-Single-CL2 and CXR-Multiple-CL2. A slight reduction in accuracy for CXR-Multiple-CL2 compared to CXR-Single-CL2 may be because of relatively more minor overfitting to the distribution of the single particular dataset from which the individual classes were coming from in CXR-Single-CL2. As expected, the results for CXR-Independent-CL2 show reduced precision and recall, with accuracy dropping from in the cross-validation results to around 88%, when using an independent test set. The observations in the experiments with CXR are consistent in CT as well. Table 3 shows the same pattern with CT-Single-CL2 and CT-Multiple-CL2 having very high accuracy compared to CT-Independent-CL2. The cross-validation results reflect the large DL models’ overfitting nature on a relatively small dataset with limited variability of the real-world scenarios. The accuracy in CT-Independent-CL2 drops from in the cross-validation results to around 79% when using the independent test set. We also notice that the accuracy with CT is lower than CXR.
5.2. Multi-class classifications: Normal, COVID, other bacterial, and viral pneumonia
Table 4 and Fig. 5 present quantitative results of the proposed CVR-Net on two different multi-class tasks: (i) -class problem for NOR vs. NCP vs. CVP (ii) -class problem for NOR vs. OBP vs. OVP vs. CVP. Similar to the binary classification, cross-validation results are reported with average and standard deviation.
Table 4.
COVID-19 recognition results from different experiments of multi-class classification (see in Table 1) applying the proposed network on CXR images employing 5-fold cross-validation.
| Different studiesa | Dataset distribution | Metrics |
||
|---|---|---|---|---|
| (TrainValTest) | Recall | Precision | Accuracy | |
| NOR: | ||||
| NCP: | ||||
| CVP: |
|
|
|
|
| CXR-Single-CL3 | Weighted Average | |||
| NOR: | ||||
| NCP: | ||||
| CVP: |
|
|
|
|
| CXR-Multiple-CL3 | Weighted Average | |||
| NOR: | ||||
| OBP: | ||||
| OVP: | ||||
| CVP: |
|
|
|
|
| CXR-Multiple-CL4 | Weighted Average | |||
X-Y-CL#: X is CXR or CT; Y denotes the way images from different sources are combined for each class during training or evaluation; CL# is the number of classes. Details in Table 1.
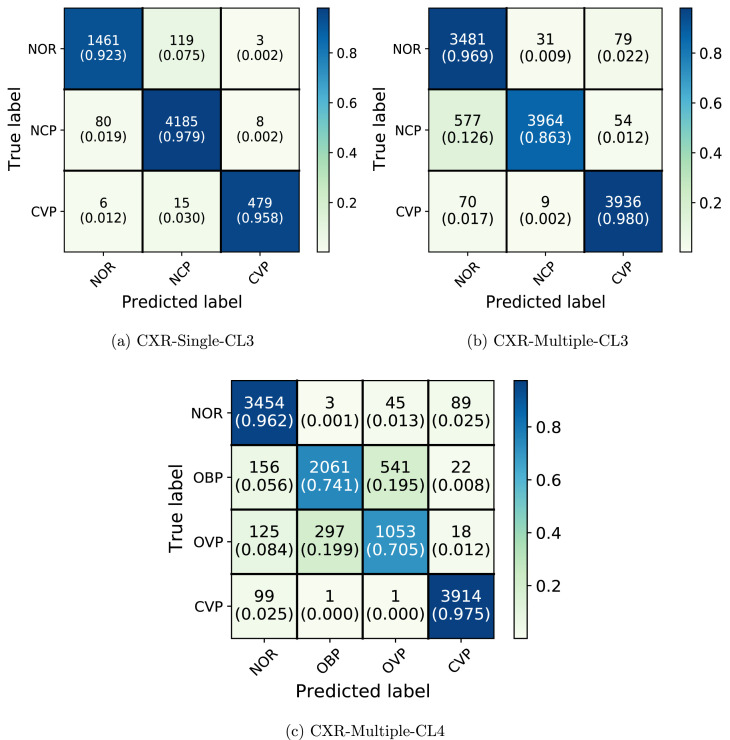
Fig. 5.
Confusion matrix for CXR-Single-CL3, CXR-Multiple-CL3, and CXR-Multiple-CL4 employing our CVR-Net.
Fig. 5 shows that in CXR-Single-CL3, NOR and NCP rarely get predicted as CVP while a small number of CVP gets predicted as NCP and NOR. Compared to CVP, a higher fraction of NOR gets predicted as NCP. This is perhaps because the NOR and NCP classes come from the same dataset source, while CVP images are from separate sources. We see that in CXR-Multiple-CL3, fractions of NOR and CVP getting predicted as NCP are much closer. It is worth noting that NOR and NCP in CXR-Multiple-CL3 have images coming from two different datasets, but these sources still do not have the CVP images coming from separate sources. It can also be observed that adding multiple data sources in NOR and NCP has substantially increased the fraction of NCP being predicted as NOR in CXR-Multiple-CL3. From Table 3, Table 4, we see that inter-fold variation is increasing with the decreased performance metrics when a new class is added with the same number of total samples when comparing CXR-Single-CL2 vs. CXR-Single-CL3 and CXR-Multiple-CL2 vs. CXR-Multiple-CL3. In CXR-Multiple-CL4, NCP is further split into other bacterial and viral Pneumonia: OBP and OVP. As seen in Fig. 5, the network confuses much more between OBP and OVP, both coming from the same dataset CXRI. Following the pattern of CXR-Single-CL3, we can also observe that nearly 14% of OBP and OVP still gets classified as NOR. CVP has relatively high precision and recall, but it is noteworthy that the source of the CVP images and the rest of the three classes do not intersect. These results further reinforce the observation in the binary classification task that seemingly high accuracy could be due to the network learning bias in the dataset design and peculiarities of individual data sources rather than the actual underlying pathology. Unlike binary classification problems, we could not evaluate with an independent test set and perform the experiments with CT scans due to the lack of publicly available datasets for these multiple classes.
5.3. Comparison to the state-of-the-art
Several recent studies report the DL models’ performance using datasets that are not publicly available [89], [90], [91]. However, we compare these methods utilizing publicly available data using the experimental setup CXR-Independent-CL2 and CT-Independent-CL2, i.e., the setup, where test set images coming from an independent dataset whose images are never used during training of the models. Table 5 manifests the performance of the proposed CVR-Net along with other widely used and state-of-the-art classification networks and COVID-19 detection networks. The hyperparameters for all the networks used in Table 5, such as learning rate, regularizations, number of epochs, optimization algorithm, etc., are described in Section 4.3 at the end. The proposed CVR-Net performs the best concerning the precision, recall, and overall accuracy in CXR and CT images. The second best is Inception-v3 for CXR and VGG-19 for CT scan.
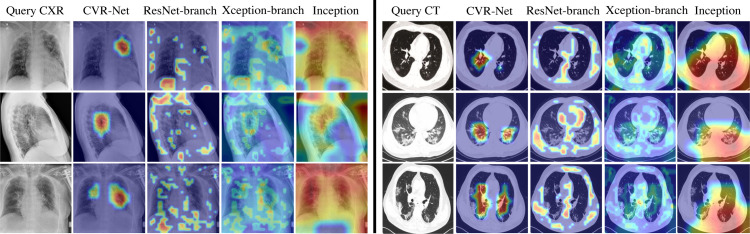
Fig. 6 visualizes the regions in the input image where the neural network is activating most of its signal from when predicting COVID-19 positive class. The activation maps are shown using GradCAM with a threshold 0.6 (maximum ) [92]. In the figure, the input images are the top three true positive images for CXR and CT, having the highest softmax prediction output for the COVID-19 class from CVR-Net. The activation map for CVR-Net as a whole is smooth and focused within the lung region, while the two branches of CVR-Net having ResNet and Xception architecture have more dispersed activation maps outside the lung region as well. This reveals that combining the two branches make the activation map more focused on the lung region. However, it is remarkably noticed that the focused region we see in the figure in the activation maps of CVR-Net does not always align with the pathology of COVID-19 seen in the CXR and CT images. For Inception, the activation maps are dispersed and smooth, but it is important to note that the images were chosen based on the highest confidence in predicting COVID-19 for CVR-Net and not for Inception.
Fig. 6.
GradCAM visualizations example, showing activation map on input query CXR and CT images of COVID-19 positive class for proposed CVR-Net, encoder-1 (ResNet), encoder-2 (Xception), and Inception.
6. Discussion and observations
We have studied the issues and challenges of DL methods on publicly available datasets for COVID-19 detection using CXRs and CT scans in this work. The results show that many current DL-based methods for COVID-19 classification over-estimate their performance. In particular, we observed two significant issues leading to such high accuracy that is likely not to translate to real-world settings: (i) the prediction classes training data come from separate individual dataset sources. This can result in the network learning the peculiarities of the dataset from which the particular class comes rather than the underlying pathology’s characteristics or features. (ii) the cross-validation results without an independent test set whose images are never used during training can overestimate the network’s performance. It is important to note that both the mentioned issues are common knowledge in machine learning but seems to have been overlooked or not emphasized enough in many recent works involving DL and COVID-19 detection [13], [34], [35], [36], [65], [80], [93], [94].
To reduce such bias and overfitting problems to some extent, we have designed an experiment where the training set contains images in each class from various dataset sources, and an independent test set is used to evaluate the deep neural networks. The results show that, as expected, the performance of the DL model reduces in this scenario. In this more realistic setting, CVR-Net performed the best when compared against other state-of-the-art classification networks. CVR-Net (architecture detailed in Section 4.2) uses multiple branches and aggregates information from different scales, creating a form of ensembling within a single network that seems to be more robust than other DL models, such as VGG, Xception, ResNet, Inception, DenseNet, and EfficientNet, as seen in Table 5. While some of the hyperparameters, such as learning rate and epochs, are adapted for each model dynamically during training, we did not exhaustively optimize the hyperparameters, regularization methods, and training protocol for each of the models separately (details in Section 4.2). For a more detailed comparison, these networks require extensive experimentation with each model to separately tune the hyperparameters and select the best regularization methods outside the scope and objective of the current paper.
-class classification task showed the model’s difficulty distinguishing bacterial Pneumonia from other viral Pneumonia. Although the results in Fig. 5 for CXR-Multiple-CL4 suggest that COVID-19 Pneumonia is well distinguished from other Pneumonia, the underlying reason is likely that these two classes come from separate data sources. To evaluate the model’s ability to distinguish different classes properly, we suggest that it is essential to have images for each class coming from the same settings, such as the same imaging protocol, machines, demography, etc. Images from multiple settings should also be included when the objective is to assess the algorithm’s ability to work in diverse settings. However, it is essential to include images from all these settings in each class in this case.
Table 5 shows higher accuracy when using CXR images compared to CT. We utilized 2D slices rather than the whole CT volume, which was not publicly available for most experimental setups. CT volume may capture details of 3D spatial information, potentially missed in these 2D slices manually selected. Thus, we cannot conclude from the results that CXR is more sensitive than CT for COVID-19 diagnosis. Moreover, the publicly available datasets come from many different sources where it is challenging to track inclusion and exclusion criteria, symptomatic vs. asymptomatic cases, and the disease severity stage at which these images were taken. Building a dataset containing these details may help identify the sensitivity of CXR vs. CT at different stages and symptom severity. This might facilitate a more informed decision for deciding between CT and CXR, which has several tradeoffs, such as patient conditions and the availability of the resource [95], [96].
7. Conclusion
This paper has explored the insights of the COVID-19 detection using the DL framework and publicly available datasets. An end-to-end DL-based model, called CVR-Net, recognizes the COVID-19 from chest radiography images with fewer false negatives. The multi-scale-multi-encoder design of the CVR-Net ensures robustness in recognition, as the final prediction probability is the aggregation of multiple scales and encoders. The experimental results show that many DL-based methods overestimate their interpretation as the data come from different individual dataset references and the cross-validation results without an independent test set. The training set from diverse sources and an independent test set can ameliorate such bias and overfitting troubles to some extent. It is also observed and suggested that it is necessary to have images for each class from identical settings like imaging protocol, machines, and demography. The results also reveal that the CXRs exhibit higher accuracy when compared to CT. We utilized 2D slices rather than the whole CT volume, unavailable for most experimental setups. CT volume may capture 3D spatial information, potentially missed in these manually selected 2D slices. However, the CXRs images can be a good choice for COVID-19 recognition as it has better performance in our experimentation, especially where CT is unavailable to collect. It can be remarked and concluded from the experiments that to accelerate the development of practical clinical DL tools, the scientific community needs to emphasize more on making publicly systematically-designed and documented datasets that have information, such as inclusion and exclusion criteria, symptomatic vs. asymptomatic cases, and the disease severity stage at which these images were taken. Future work will improve the performance by segmenting the lung and adding more distinctive training samples to all the classes. We also intend to deploy our trained CVR-Net to a web application for clinical utilization.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
We thank the people who contributed to making the COVID-19 related radiography images public. We also thank radiologist Dr. Ram Kumar Ghimire, for feedback on the characteristics of COVID-19 seen in specific CXR and CT images.
Footnotes
Data and code availability
Source code and trained model available at https://github.com/kamruleee51/CVR-Net.
References
- 1.World Health Organization . 2020. Naming the coronavirus disease (COVID-19) https://tinyurl.com/25r7muwv [Accessed: 16 December 2021] [Google Scholar]
- 2.World Health Organization . 2022. WHO Coronavirus disease (COVID-19) dashboard. https://covid19.who.int/ [Accessed: 12 December 2021] [Google Scholar]
- 3.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., et al. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang T., Wang Y.-C., Shen C.-F., Cheng C.-M. Multidisciplinary Digital Publishing Institute; 2020. Point-of-care RNA-based diagnostic device for COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.NEWS A.J. 2020. Bangladesh Scientists create kit. Can it help detect COVID-19? https://bit.ly/aj2020corona [Accessed: 19 December 2021] [Google Scholar]
- 9.COVID C. ArcGIS. johns hopkins CSSE. retrieved april, Vol. 8. 2020. Global cases by the center for systems science and engineering (CSSE) at johns hopkins university (JHU) p. 19. [Google Scholar]
- 10.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu X., Jiang X., Ma C., Du P., Li X., Lv S., et al. A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering. 2020 doi: 10.1016/j.eng.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh D., Kumar V., Kaur M. Classification of COVID-19 patients from chest CT images using multi-objective differential evolution–based convolutional neural networks. Eur J Clin Microbiol Infect Diseases. 2020:1–11. doi: 10.1007/s10096-020-03901-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee E.Y., Ng M.-Y., Khong P.-L. COVID-19 pneumonia: what has CT taught us? Lancet Infect Diseases. 2020;20(4):384–385. doi: 10.1016/S1473-3099(20)30134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 16.Krizhevsky A., Sutskever I., Hinton G.E. Advances in neural information processing systems. 2012. Imagenet classification with deep convolutional neural networks; pp. 1097–1105. [Google Scholar]
- 17.Yıldırım O., Pławiak P., Tan R.-S., Acharya U.R. Arrhythmia detection using deep convolutional neural network with long duration ECG signals. Comput Biol Med. 2018;102:411–420. doi: 10.1016/j.compbiomed.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 18.Hannun A.Y., Rajpurkar P., Haghpanahi M., Tison G.H., Bourn C., Turakhia M.P., et al. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat Med. 2019;25(1):65. doi: 10.1038/s41591-018-0268-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Acharya U.R., Oh S.L., Hagiwara Y., Tan J.H., Adam M., Gertych A., et al. A deep convolutional neural network model to classify heartbeats. Comput Biol Med. 2017;89:389–396. doi: 10.1016/j.compbiomed.2017.08.022. [DOI] [PubMed] [Google Scholar]
- 20.Hasan M.K., Dahal L., Samarakoon P.N., Tushar F.I., Martí R. DSNet: Automatic dermoscopic skin lesion segmentation. Comput Biol Med. 2020;120 doi: 10.1016/j.compbiomed.2020.103738. [DOI] [PubMed] [Google Scholar]
- 21.Esteva A., Kuprel B., Novoa R.A., Ko J., Swetter S.M., Blau H.M., et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Codella N.C., Nguyen Q.-B., Pankanti S., Gutman D.A., Helba B., Halpern A.C., Smith J.R. Deep learning ensembles for melanoma recognition in dermoscopy images. IBM J Res Dev. 2017;61(4/5) 5–1. [Google Scholar]
- 23.Hasan M.K., Elahi M.T.E., Alam M.A., Jawad M.T., Martí R. DermoExpert: Skin lesion classification using a hybrid convolutional neural network through segmentation, transfer learning, and augmentation. Inform Med Unlock. 2022 [Google Scholar]
- 24.Hasan M.K., Roy S., Mondal C., Alam M.A., Elahi M.T.E., Dutta A., et al. Dermo-DOCTOR: A framework for concurrent skin lesion detection and recognition using a deep convolutional neural network with end-to-end dual encoders. Biomed Signal Process Control. 2021;68 [Google Scholar]
- 25.Celik Y., Talo M., Yildirim O., Karabatak M., Acharya U.R. Automated invasive ductal carcinoma detection based using deep transfer learning with whole-slide images. Pattern Recognit Lett. 2020 [Google Scholar]
- 26.Cruz-Roa A., Basavanhally A., González F., Gilmore H., Feldman M., Ganesan S., et al. Medical imaging 2014: digital pathology, Vol. 9041. International Society for Optics and Photonics; 2014. Automatic detection of invasive ductal carcinoma in whole slide images with convolutional neural networks. [Google Scholar]
- 27.Hasan M.K., Aleef T.A., Roy S. 2020 IEEE region 10 symposium. IEEE; 2020. Automatic mass classification in breast using transfer learning of deep convolutional neural network and support vector machine; pp. 110–113. [Google Scholar]
- 28.Talo M., Yildirim O., Baloglu U.B., Aydin G., Acharya U.R. Convolutional neural networks for multi-class brain disease detection using MRI images. Comput Med Imaging Graph. 2019;78 doi: 10.1016/j.compmedimag.2019.101673. [DOI] [PubMed] [Google Scholar]
- 29.Rajpurkar P., Irvin J., Zhu K., Yang B., Mehta H., Duan T., et al. 2017. Chexnet: Radiologist-level pneumonia detection on chest x-rays with deep learning. ArXiv:1711.05225. [Google Scholar]
- 30.Tan J.H., Fujita H., Sivaprasad S., Bhandary S.V., Rao A.K., Chua K.C., et al. Automated segmentation of exudates, haemorrhages, microaneurysms using single convolutional neural network. Inform Sci. 2017;420:66–76. [Google Scholar]
- 31.Hasan M.K., Alam M.A., Elahi M.T.E., Roy S., Martí R. DRNet: Segmentation and localization of optic disc and fovea from diabetic retinopathy image. Artif Intell Med. 2021;111 doi: 10.1016/j.artmed.2020.102001. [DOI] [PubMed] [Google Scholar]
- 32.Hasan M.K., Calvet L., Rabbani N., Bartoli A. Detection, segmentation, and 3D pose estimation of surgical tools using convolutional neural networks and algebraic geometry. Med Image Anal. 2021;70 doi: 10.1016/j.media.2021.101994. [DOI] [PubMed] [Google Scholar]
- 33.Gaál G., Maga B., Lukács A. 2020. Attention u-net based adversarial architectures for chest x-ray lung segmentation. ArXiv:2003.10304. [Google Scholar]
- 34.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. 2020 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khan A.I., Shah J.L., Bhat M.M. Coronet: A deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput Methods Programs Biomed. 2020 doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Narin A., Kaya C., Pamuk Z. 2020. Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. ArXiv:2003.10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghoshal B., Tucker A. 2020. Estimating uncertainty and interpretability in deep learning for coronavirus (COVID-19) detection. ArXiv:2003.10769. [Google Scholar]
- 38.Abbas A., Abdelsamea M.M., Gaber M.M. 2020. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. ArXiv:2003.13815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abbas A., Abdelsamea M.M., Gaber M.M. Detrac: Transfer learning of class decomposed medical images in convolutional neural networks. IEEE Access. 2020;8:74901–74913. [Google Scholar]
- 40.Zhao J., Zhang Y., He X., Xie P. 2020. COVID-CT-dataset: a CT scan dataset about COVID-19. ArXiv:2003.13865. [Google Scholar]
- 41.Afshar P., Heidarian S., Naderkhani F., Oikonomou A., Plataniotis K.N., Mohammadi A. 2020. Covid-caps: A capsule network-based framework for identification of covid-19 cases from x-ray images. ArXiv:2004.02696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ioffe S., Szegedy C. 2015. Batch normalization: Accelerating deep network training by reducing internal covariate shift. ArXiv:1502.03167. [Google Scholar]
- 43.He X., Wang S., Shi S., Chu X., Tang J., Liu X., et al. Cold Spring Harbor Laboratory Press; 2020. Benchmarking deep learning models and automated model design for COVID-19 detection with chest CT scans. MedRxiv. [Google Scholar]
- 44.Farooq M., Hafeez A. 2020. Covid-resnet: A deep learning framework for screening of covid19 from radiographs. ArXiv:2003.14395. [Google Scholar]
- 45.Hasan M.K., Jawad M.T., Hasan K.N.I., Partha S.B., Al Masba M.M., Saha S., et al. COVID-19 identification from volumetric chest CT scans using a progressively resized 3D-CNN incorporating segmentation, augmentation, and class-rebalancing. Inform Med Unlock. 2021;26 doi: 10.1016/j.imu.2021.100709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ozkaya U., Ozturk S., Barstugan M. 2020. Coronavirus (COVID-19) classification using deep features fusion and ranking technique. ArXiv:2004.03698. [Google Scholar]
- 47.Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, et al. Going deeper with convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition. 2015, p. 1–9.
- 48.Furey T.S., Cristianini N., Duffy N., Bednarski D.W., Schummer M., Haussler D. Support vector machine classification and validation of cancer tissue samples using microarray expression data. Bioinformatics. 2000;16(10):906–914. doi: 10.1093/bioinformatics/16.10.906. [DOI] [PubMed] [Google Scholar]
- 49.Zhou N., Wang L. A modified T-test feature selection method and its application on the HapMap genotype data. Genom Proteom Bioinform. 2007;5(3–4):242–249. doi: 10.1016/S1672-0229(08)60011-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hasan M.K., Jawad M.T., Dutta A., Awal M.A., Islam M.A., Masud M., et al. Associating measles vaccine uptake classification and its underlying factors using an ensemble of machine learning models. IEEE Access. 2021;9:119613–119628. [Google Scholar]
- 51.Rajaraman S., Siegelman J., Alderson P.O., Folio L.S., Folio L.R., Antani S.K. 2020. Iteratively pruned deep learning ensembles for COVID-19 detection in chest X-rays. ArXiv:2004.08379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pham H., Guan M.Y., Zoph B., Le Q.V., Dean J. 2018. Efficient neural architecture search via parameter sharing. ArXiv:1802.03268. [Google Scholar]
- 53.Bergstra J., Bengio Y. Random search for hyper-parameter optimization. J Mach Learn Res. 2012;13(1):281–305. [Google Scholar]
- 54.Toğaçar M., Ergen B., Cömert Z. COVID-19 detection using deep learning models to exploit social mimic optimization and structured chest X-ray images using fuzzy color and stacking approaches. Comput Biol Med. 2020 doi: 10.1016/j.compbiomed.2020.103805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Balochian S., Baloochian H. Social mimic optimization algorithm and engineering applications. Expert Syst Appl. 2019;134:178–191. [Google Scholar]
- 56.Khan M.A., Kadry S., Zhang Y.-D., Akram T., Sharif M., Rehman A., et al. Prediction of COVID-19-pneumonia based on selected deep features and one class kernel extreme learning machine. Comput Electr Eng. 2021;90 doi: 10.1016/j.compeleceng.2020.106960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Narin A., Kaya C., Pamuk Z. Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. Pattern Anal Appl. 2021;24(3):1207–1220. doi: 10.1007/s10044-021-00984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sedik A., Hammad M., El-Samie A., Fathi E., Gupta B.B., El-Latif A., et al. Efficient deep learning approach for augmented detection of coronavirus disease. Neural Comput Appl. 2021:1–18. doi: 10.1007/s00521-020-05410-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanida T., Sideris A., Tsiktsiris D., Dasygenis M. Lightweight neural network for COVID-19 detection from chest X-ray images implemented on an embedded system. Technologies. 2022;10(2):37. [Google Scholar]
- 60.Turkoglu M. COVIDetectioNet: COVID-19 diagnosis system based on X-ray images using features selected from pre-learned deep features ensemble. Appl Intell. 2021;51(3):1213–1226. doi: 10.1007/s10489-020-01888-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.ElAraby M.E., Elzeki O.M., Shams M.Y., Mahmoud A., Salem H. A novel gray-scale spatial exploitation learning net for COVID-19 by crawling internet resources. Biomed Signal Process Control. 2022;73 doi: 10.1016/j.bspc.2021.103441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Monday H.N., Li J., Nneji G.U., Nahar S., Hossin M.A., Jackson J. Healthcare, Vol. 10. MDPI; 2022. COVID-19 pneumonia classification based on NeuroWavelet capsule network; p. 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sakthivel R., Thaseen I.S., Vanitha M., Deepa M., Angulakshmi M., Mangayarkarasi R., et al. An efficient hardware architecture based on an ensemble of deep learning models for COVID-19 prediction. Sustainable Cities Soc. 2022 doi: 10.1016/j.scs.2022.103713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Apostolopoulos I.D., Aznaouridis S.I., Tzani M.A. Extracting possibly representative COVID-19 biomarkers from X-Ray images with deep learning approach and image data related to pulmonary diseases. J Med Biol Eng. 2020:1. doi: 10.1007/s40846-020-00529-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020:1. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hall L.O., Paul R., Goldgof D.B., Goldgof G.M. 2020. Finding covid-19 from chest x-rays using deep learning on a small dataset. ArXiv:2004.02060. [Google Scholar]
- 67.Huang L., Han R., Ai T., Yu P., Kang H., Tao Q., et al. Serial quantitative chest ct assessment of covid-19: Deep-learning approach. Radiol: Cardiothoracic Imaging. 2020;2(2) doi: 10.1148/ryct.2020200075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mahmud T., Rahman M.A., Fattah S.A. CovXNet: A multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization. Comput Biol Med. 2020;122 doi: 10.1016/j.compbiomed.2020.103869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Minaee S., Kafieh R., Sonka M., Yazdani S., Jamalipour Soufi G. Deep-COVID: Predicting COVID-19 from chest X-ray images using deep transfer learning. Med Image Anal. 2020;65 doi: 10.1016/j.media.2020.101794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oh Y., Park S., Ye J.C. Deep learning covid-19 features on cxr using limited training data sets. IEEE Trans Med Imaging. 2020 doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- 71.Zhang K., Liu X., Shen J., Li Z., Sang Y., Wu X., et al. Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of covid-19 pneumonia using computed tomography. Cell. 2020 doi: 10.1016/j.cell.2020.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mooney P. 2018. Chest X-Ray images (pneumonia) https://tinyurl.com/33sjpfz7 [Accessed: 22 December 2021] [Google Scholar]
- 73.Cohen J.P., Morrison P., Dao L., Roth K., Duong T.Q., Ghassemi M. 2020. COVID-19 image data collection: Prospective predictions are the future. ArXiv:2006.11988. [Google Scholar]
- 74.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM. Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: Proceedings of the IEEE conference on computer vision and pattern recognition. 2017, p. 2097–106.
- 75.Chowdhury M.E., Rahman T., Khandakar A., Mazhar R., Kadir M.A., Mahbub Z.B., et al. 2020. Can AI help in screening viral and COVID-19 pneumonia? arXiv preprint arXiv:2003.13145. [Google Scholar]
- 76.Bustos A., Pertusa A., Salinas J.-M., de la Iglesia-Vayá M. Padchest: A large chest x-ray image dataset with multi-label annotated reports. Med Image Anal. 2020;66 doi: 10.1016/j.media.2020.101797. [DOI] [PubMed] [Google Scholar]
- 77.Irvin J., Rajpurkar P., Ko M., Yu Y., Ciurea-Ilcus S., Chute C., et al. Proceedings of the AAAI conference on artificial intelligence, Vol. 33. 2019. Chexpert: A large chest radiograph dataset with uncertainty labels and expert comparison; pp. 590–597. [Google Scholar]
- 78.Angelov P., Almeida E. 2020. Explainable-by-design approach for covid-19 classification via ct-scan. MedRxiv. [Google Scholar]
- 79.Ning W., Lei S., Yang J., Cao Y., Jiang P., Yang Q., et al. 2020. iCTCF: an integrative resource of chest computed tomography images and clinical features of patients with COVID-19 pneumonia. [Google Scholar]
- 80.Wang L., Wong A. 2020. COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-Ray images. arXiv preprint arXiv:2003.09871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition. 2016, p. 770–8.
- 82.Chollet F. Xception: Deep learning with depthwise separable convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition. 2017, p. 1251–8.
- 83.Lin M., Chen Q., Yan S. 2013. Network in network. ArXiv:1312.4400. [Google Scholar]
- 84.Zhang Z., Sabuncu M. Advances in neural information processing systems. 2018. Generalized cross entropy loss for training deep neural networks with noisy labels; pp. 8778–8788. [Google Scholar]
- 85.Shin H.-C., Roth H.R., Gao M., Lu L., Xu Z., Nogues I., et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging. 2016;35(5):1285–1298. doi: 10.1109/TMI.2016.2528162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kingma D.P., Ba J. 2014. Adam: A method for stochastic optimization. ArXiv:1412.6980. [Google Scholar]
- 87.Chollet F. 2015. Keras. GitHub Repository,GitHub, https://github.com/fchollet/keras. [Google Scholar]
- 88.Hasan M.K., Alam M.A., Roy S., Dutta A., Jawad M.T., Das S. Missing value imputation affects the performance of machine learning: A review and analysis of the literature (2010–2021) Inform Med Unlock. 2021;27 [Google Scholar]
- 89.Harmon S.A., Sanford T.H., Xu S., Turkbey E.B., Roth H., Xu Z., et al. Artificial intelligence for the detection of COVID-19 pneumonia on chest CT using multinational datasets. Nature Commun. 2020;11(1):1–7. doi: 10.1038/s41467-020-17971-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang Z., Xiao Y., Li Y., Zhang J., Lu F., Hou M., et al. Automatically discriminating and localizing COVID-19 from community-acquired pneumonia on chest X-rays. Pattern Recognit. 2020 doi: 10.1016/j.patcog.2020.107613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Song Y., Zheng S., Li L., Zhang X., Zhang X., Huang Z., et al. Cold Spring Harbor Laboratory Press; 2020. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. MedRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In: Proceedings of the IEEE international conference on computer vision. 2017, p. 618–26.
- 93.Hemdan E.E.-D., Shouman M.A., Karar M.E. 2020. Covidx-net: A framework of deep learning classifiers to diagnose covid-19 in x-ray images. ArXiv:2003.11055. [Google Scholar]
- 94.Sethy P.K., Behera S.K. Preprints; 2020. Detection of coronavirus disease (covid-19) based on deep features, Vol. 2020030300; p. 2020. [Google Scholar]
- 95.Cleverley J., Piper J., Jones M.M. The role of chest radiography in confirming covid-19 pneumonia. Bmj. 2020;370 doi: 10.1136/bmj.m2426. [DOI] [PubMed] [Google Scholar]
- 96.Rubin G.D., Ryerson C.J., Haramati L.B., Sverzellati N., Kanne J.P., Raoof S., et al. The role of chest imaging in patient management during the COVID-19 pandemic: a multinational consensus statement from the fleischner society. Chest. 2020 doi: 10.1016/j.chest.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Source code and trained model available at https://github.com/kamruleee51/CVR-Net.