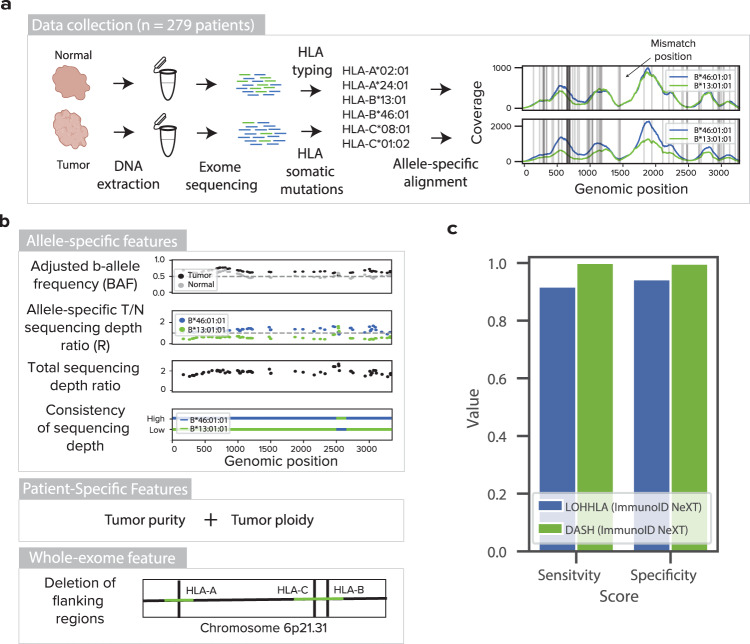
Fig. 1. Training, performance, and limit of detection of DASH algorithm.
a Tumor and adjacent normal samples were collected for 279 patients. DNA was extracted and exome sequencing was performed. HLA typing was derived from the normal sample and HLA somatic mutations were obtained from the tumor sample. Reads mapping to an HLA reference database were assembled and mapped onto the patient-specific HLA alleles. Horizontal gray lines denote positions where the homologous alleles differ from each other. b Features used to train the DASH algorithm include the adjusted b-allele frequency, sequencing depth ratio, total sequencing depth ratio, consistency of sequencing depth, tumor purity, tumor ploidy and deletion of flanking regions. Dashed gray lines represent the expected values for a sample without copy number change. All features were used to train an XGBoost prediction model. c A bar plot showing the sensitivity and specificity of HLA LOH detection by DASH and LOHHLA for samples in the 10-fold cross validation data set. Both algorithms were tested on the HLA-enhanced ImmunoID NeXT Platform. Only samples with at least 20% tumor purity were included. Source data are provided with this paper.