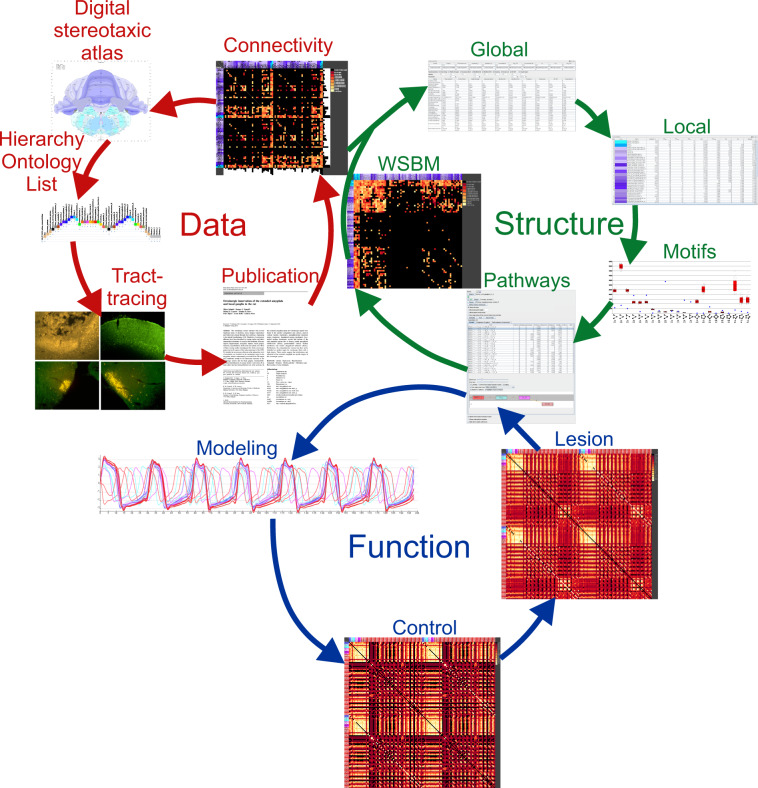
Fig. 1.
Schematic outline of the brainstem connectome generation and simulations in the neuroVIISAS framework. Data generation starts with hypothesis of possible connections using stereotaxic atlases and concepts of knowledge (ontologies). Then TT experiments are performed and connections are published. Original research publications were evaluated and connections described herein were collated and imported into the rat connectome database to build the connectivity of adjacency matrices. These are starting points for global and local as well as motif network analysis. neuroVIISAS provides tools to investigate pathways and community detection algorithms like weighted stochastic block matching (WSBM), Louvain modularity and spectral graph analysis among oth-ers. The weights of a pathway can be reduced to model demyelination disorders (lesion) which can be compared with control coactivation matrices derived from excitatory FitzHugh Nagumo network propagation models.