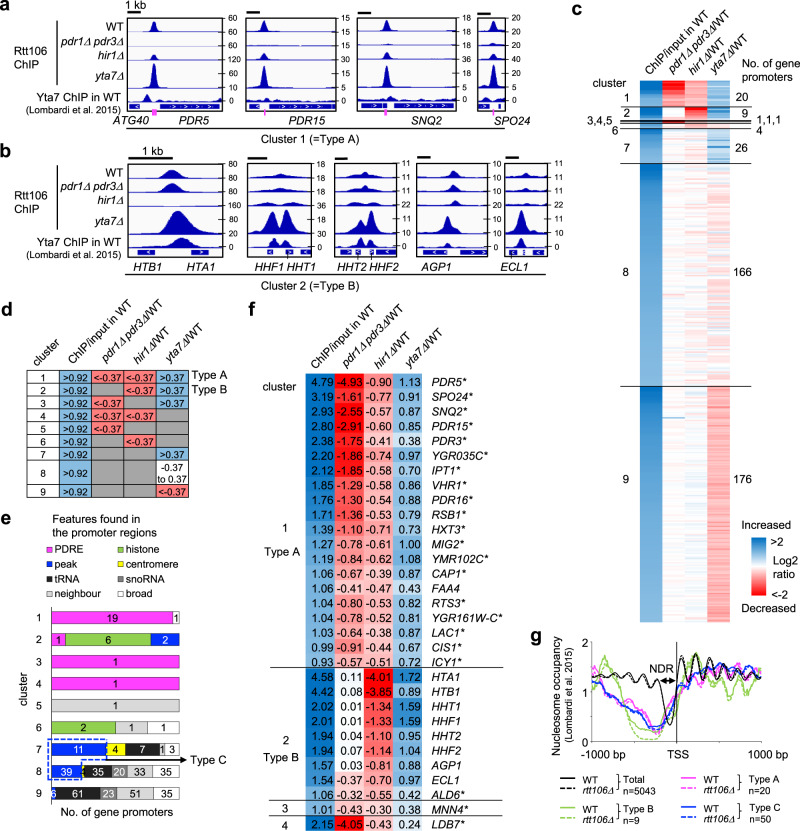
Fig. 2. A series of ChIP-seq tests reveal three distinct modes of Rtt106 binding to promoters by clustering analysis.
a Specimen views of promoters where Rtt106 binding is dependent on Pdr1/3 and Hir1 and limited by Yta7 (falling into cluster 1, designated as Type A). Yta7 binding tested by ChIP-seq analysis29 is shown alongside Rtt106 binding. Positions of PDRE indicated by magenta lines. b Specimen views of promoters where Rtt106 binding is independent of Pdr1/3, dependent on Hir1 and limited by Yta7 (falling into cluster 2, designated as Type B). ChIP-seq analyses of Rtt106 in WT, pdr1Δ pdr3Δ, hir1Δ and yta7Δ mutants are shown. c Clustering analysis of Rtt106 promoter association based on Rtt106 binding in WT and its changes in pdr1Δ pdr3Δ, hir1Δ and yta7Δ. Log2 fold enrichment of Rtt106 binding in WT at promoters (ChIP/input in WT) and log2 fold changes of Rtt106 binding in pdr1Δ pdr3Δ, hir1Δ and yta7Δ compared to that in WT (pdr1Δ pdr3Δ/WT, hir1Δ/WT and yta7Δ/WT, respectively) were calculated as described in Methods. d Criteria used for clustering in panel c. e Features found in promoters categorised into clusters 1–9. Numbers of gene promoters containing PDRE (PDR responsive element; magenta), centromere (yellow), tRNA (black) or snoRNA (dark grey), and those containing Rtt106 signals leaked from highly expressing neighbour genes (‘neighbour’; light grey) are shown. Histone gene promoters (‘histone’; green) were assessed separately. Other than the above features, numbers of promoters with Rtt106 peaks (‘peak’; blue) or broad signals (‘broad’; white) are shown. Promoters with ‘peaks’ in clusters 7 and 8 designated as Type C. f Gene promoters categorised into clusters 1–4. Log2 changes are shown as in c. Asterisks indicate gene promoters containing PDRE. g Nucleosome occupancy analysed by MNase-seq in WT and rtt106Δ (Lombardi et al. 201529) aligned to transcription start site (TSS). Nucleosome-depleted region (NDR) in average of all gene promoters is indicated.